Template class that stores 3D voxel data. More...
#include <voxels.h>
Public Member Functions | |
| Voxels () | |
| Constructor. More... | |
| ~Voxels () | |
| Destructor. More... | |
| void | swap (Voxels< T > &v) |
| Swap object contents with other voxel object. More... | |
| void | copy (Voxels< T > &newCopy) const |
| Clone data into another object. More... | |
| void | setPoint (const AtomProbe::Point3D &pt, const T &val) |
| Set the value of a point in the dataset. More... | |
| T | getPointData (const AtomProbe::Point3D &pt) const |
| Retrieve the value of a datapoint, this is rounded to the nearest voxel. More... | |
| void | getIndex (size_t &x, size_t &y, size_t &z, const AtomProbe::Point3D &p) const |
| Retrieve the XYZ voxel location associated with a given position. More... | |
| void | getIndexWithUpper (size_t &x, size_t &y, size_t &z, const AtomProbe::Point3D &p) const |
| Retrieve the XYZ voxel location associated with a given position,. More... | |
| AtomProbe::Point3D | getPoint (size_t x, size_t y, size_t z) const |
| Get the position associated with an XYZ voxel. More... | |
| T | getData (size_t x, size_t y, size_t z) const |
| Retrieve the value of a specific voxel. More... | |
| T | getData (size_t i) const |
| Retrieve value of the nth voxel. More... | |
| void | setEntry (size_t n, const T &val) |
| void | setData (size_t x, size_t y, size_t z, const T &val) |
| Retrieve a reference to the data ata given position. More... | |
| void | setData (size_t n, const T &val) |
| Set the value of nth point in the dataset. More... | |
| void | getInterpolatedData (const AtomProbe::Point3D &pt, T &v) const |
| void | getInterpSlice (size_t normal, float offset, T *p, size_t interpMode=VOX_INTERP_NONE) const |
| void | getSlice (size_t normal, size_t offset, T *p) const |
| void | getSize (size_t &x, size_t &y, size_t &z) const |
| Get the size of the data field. More... | |
| size_t | resize (size_t newX, size_t newY, size_t newZ, const AtomProbe::Point3D &newMinBound=AtomProbe::Point3D(0.0f, 0.0f, 0.0f), const AtomProbe::Point3D &newMaxBound=AtomProbe::Point3D(1.0f, 1.0f, 1.0f)) |
| Resize the data field. More... | |
| size_t | resize (const Voxels< T > &v) |
| size_t | resizeKeepData (size_t newX, size_t newY, size_t newZ, unsigned int direction=CLIP_LOWER_SOUTH_WEST, const AtomProbe::Point3D &newMinBound=AtomProbe::Point3D(0.0f, 0.0f, 0.0f), const AtomProbe::Point3D &newMaxBound=AtomProbe::Point3D(1.0f, 1.0f, 1.0f), const T &fill=T(0), bool doFill=false) |
| Resize the data field, maintaining data as best as possible. More... | |
| size_t | deprecatedGetEdgeUniqueIndex (size_t x, size_t y, size_t z, unsigned int edge) const |
| DEPRECATED FUNCTION : Get a unique integer that corresponds to the edge index for the voxel; where edges are shared between voxels. More... | |
| size_t | getCellUniqueEdgeIndex (size_t x, size_t y, size_t z, unsigned int edge) const |
| Get a unique integer that corresponds to an edge index for the voxel; where edges are shared between voxels. More... | |
| void | getEdgeEnds (size_t edgeIndex, AtomProbe::Point3D &a, AtomProbe::Point3D &b) const |
| Convert the edge index (as generated by getEdgeUniqueIndex) into a cenre position. More... | |
| void | getEdgeCell (size_t edgeUniqId, size_t &x, size_t &y, size_t &z, size_t &axis) const |
| Convert edge index (only as generted by getCellUniqueEdgeIndex) into a cell & axis value. More... | |
| void | getEdgeEndApproxVals (size_t edgeUniqId, T &a, T &b) const |
| Return the values that are associated with the edge ends, as returned by getEdgeEnds. More... | |
| T | getSum (const T &initialVal=T(0.0)) const |
| Get the total value of the data field. More... | |
| size_t | count (const T &minIntensity) const |
| count the number of cells with at least this intensity More... | |
| void | fill (const T &val) |
| Fill all voxels with a given value. More... | |
| AtomProbe::Point3D | getMinBounds () const |
| Get the bounding box vertex (min/max) More... | |
| AtomProbe::Point3D | getMaxBounds () const |
| void | getAxisBounds (size_t axis, float &minV, float &maxV) const |
| AtomProbe::Point3D | getPitch () const |
| ! Get the spacing for a unit cell More... | |
| void | setBounds (const AtomProbe::Point3D &pMin, const AtomProbe::Point3D &pMax) |
| Set the bounding size. More... | |
| void | setBounds (const BoundCube &b) |
| void | getBounds (AtomProbe::Point3D &pMin, AtomProbe::Point3D &pMax) const |
| Get the bounding size. More... | |
| void | getBounds (BoundCube &bc) const |
| size_t | init (size_t nX, size_t nY, size_t nZ, const BoundCube &bound) |
| Initialise the voxel storage. More... | |
| size_t | init (size_t nX, size_t nY, size_t nZ) |
| Initialise the voxel storage. More... | |
| size_t | loadFile (const char *cpFilename, size_t nX, size_t nY, size_t nZ, bool silent=false) |
| Load the voxels from file. More... | |
| size_t | writeFile (const char *cpFilename) const |
| Write the voxel objects in column major written out to file. More... | |
| void | threshold (const T &thresh, bool keepUpper, const T &newVal) |
| Threshold the voxels, keeping either the lower or upper values above this threshold. More... | |
| void | thresholdToBoolMask (const T &thresh, bool keepUpper, Voxels< bool > &result) const |
| Generate a boolean voxel field stating whether the data is above or below the threshold specified. More... | |
| void | applyMask (const Voxels< bool > &mask, const T &newVal, bool invert=false) |
| Apply a mask to this dataset, where the mask is true (if not inverting), replace with specified val (newVal) More... | |
| void | thresholdForPosition (std::vector< AtomProbe::Point3D > &p, const T &thresh, bool lowerEq=false) const |
| Find the positions of the voxels that are above or below a given value. More... | |
| void | binarise (Voxels< T > &result, const T &thresh, const T &onThresh, const T &offThresh) const |
| Binarise the data into a result vector. More... | |
| void | clear () |
| Empty the data. More... | |
| T | min () const |
| Find minimum in dataset. More... | |
| T | max () const |
| Find maximum in dataset. More... | |
| void | minMax (T &min, T &max) const |
| Find both min and max in dataset in the same loop. More... | |
| int | countPoints (const std::vector< AtomProbe::Point3D > &points, bool noWrap=true, bool doErase=true) |
| Generate a dataset that consists of the counts of points in a given voxel. More... | |
| int | countIons (const std::vector< AtomProbe::IonHit > &ions, bool noWrap=true, bool doErase=true) |
| Generate a dataset that consists of the counts of points in a given voxel. More... | |
| T | trapezIntegral () const |
| Integrate the datataset via the trapezoidal method. More... | |
| void | calculateDensity () |
| Convert voxel intensity into voxel density. More... | |
| float | getBinVolume () const |
| void | operator/= (const Voxels< T > &v) |
| Element wise division. More... | |
| void | operator/= (const T &v) |
| bool | operator== (const Voxels< T > &v) const |
| size_t | size () const |
Static Public Member Functions | |
| static size_t | sizeofType () |
| Return the sizeof value for the T type. More... | |
Detailed Description
template<class T>
class AtomProbe::Voxels< T >
Template class that stores 3D voxel data.
To instantiate this class, objects must have basic mathematical operators, such as * + - and =
Constructor & Destructor Documentation
◆ Voxels()
| AtomProbe::Voxels< T >::Voxels | ( | ) |
◆ ~Voxels()
| AtomProbe::Voxels< T >::~Voxels | ( | ) |
Member Function Documentation
◆ applyMask()
| void AtomProbe::Voxels< T >::applyMask | ( | const Voxels< bool > & | mask, |
| const T & | newVal, | ||
| bool | invert = false |
||
| ) |
Apply a mask to this dataset, where the mask is true (if not inverting), replace with specified val (newVal)
Definition at line 1297 of file voxels.h.
References ASSERT, AtomProbe::Voxels< T >::getData(), AtomProbe::Voxels< T >::getMaxBounds(), AtomProbe::Voxels< T >::getMinBounds(), AtomProbe::Voxels< T >::getSize(), and AtomProbe::Voxels< T >::size().
Referenced by AtomProbe::Voxels< T >::getBounds().
◆ binarise()
| void AtomProbe::Voxels< T >::binarise | ( | Voxels< T > & | result, |
| const T & | thresh, | ||
| const T & | onThresh, | ||
| const T & | offThresh | ||
| ) | const |
Binarise the data into a result vector.
Definition at line 1330 of file voxels.h.
References AtomProbe::Voxels< T >::getData(), AtomProbe::Voxels< T >::resize(), and AtomProbe::Voxels< T >::setData().
Referenced by AtomProbe::Voxels< T >::sizeofType().
◆ calculateDensity()
| void AtomProbe::Voxels< T >::calculateDensity | ( | ) |
Convert voxel intensity into voxel density.
Definition at line 1469 of file voxels.h.
References AtomProbe::Voxels< T >::size().
Referenced by AtomProbe::Voxels< T >::clear().
◆ clear()
|
inline |
Empty the data.
Definition at line 314 of file voxels.h.
References AtomProbe::Voxels< T >::calculateDensity(), AtomProbe::Voxels< T >::countIons(), AtomProbe::Voxels< T >::countPoints(), AtomProbe::Voxels< T >::getBinVolume(), AtomProbe::Voxels< T >::max(), AtomProbe::Voxels< T >::min(), AtomProbe::Voxels< T >::minMax(), AtomProbe::Voxels< T >::operator/=(), AtomProbe::Voxels< T >::operator==(), and AtomProbe::Voxels< T >::trapezIntegral().
◆ copy()
| void AtomProbe::Voxels< T >::copy | ( | Voxels< T > & | newCopy | ) | const |
◆ count()
| size_t AtomProbe::Voxels< T >::count | ( | const T & | minIntensity | ) | const |
count the number of cells with at least this intensity
Definition at line 1156 of file voxels.h.
Referenced by AtomProbe::Voxels< T >::setEntry().
◆ countIons()
| int AtomProbe::Voxels< T >::countIons | ( | const std::vector< AtomProbe::IonHit > & | ions, |
| bool | noWrap = true, |
||
| bool | doErase = true |
||
| ) |
Generate a dataset that consists of the counts of points in a given voxel.
Ensure that the voxel scaling factors are set by calling ::setBounds() with the appropriate parameters for your data. Disabling nowrap allows you to "saturate" your data field in the case of dense regions, rather than let wrap-around errors occur
Definition at line 1418 of file voxels.h.
References AtomProbe::Voxels< T >::fill(), AtomProbe::Voxels< T >::getData(), AtomProbe::Voxels< T >::getIndex(), and AtomProbe::Voxels< T >::setData().
Referenced by AtomProbe::Voxels< T >::clear().
◆ countPoints()
| int AtomProbe::Voxels< T >::countPoints | ( | const std::vector< AtomProbe::Point3D > & | points, |
| bool | noWrap = true, |
||
| bool | doErase = true |
||
| ) |
Generate a dataset that consists of the counts of points in a given voxel.
Ensure that the voxel scaling factors are set by calling ::setBounds() with the appropriate parameters for your data. Disabling nowrap allows you to "saturate" your data field in the case of dense regions, rather than let wrap-around errors occur
Definition at line 1382 of file voxels.h.
References AtomProbe::Voxels< T >::fill(), AtomProbe::Voxels< T >::getData(), AtomProbe::Voxels< T >::getIndex(), and AtomProbe::Voxels< T >::setData().
Referenced by AtomProbe::Voxels< T >::clear().
◆ deprecatedGetEdgeUniqueIndex()
| size_t AtomProbe::Voxels< T >::deprecatedGetEdgeUniqueIndex | ( | size_t | x, |
| size_t | y, | ||
| size_t | z, | ||
| unsigned int | edge | ||
| ) | const |
DEPRECATED FUNCTION : Get a unique integer that corresponds to the edge index for the voxel; where edges are shared between voxels.
Each voxel has 12 edges. These are shared (except voxels that on zero or positive boundary). Return a unique index that corresponds to a specified edge (0->11). Index CANNOT be inverted to yield cell
Definition at line 497 of file voxels.h.
Referenced by AtomProbe::marchingCubes(), and AtomProbe::Voxels< T >::setEntry().
◆ fill()
| void AtomProbe::Voxels< T >::fill | ( | const T & | val | ) |
Fill all voxels with a given value.
Definition at line 1536 of file voxels.h.
Referenced by AtomProbe::Voxels< T >::countIons(), AtomProbe::Voxels< T >::countPoints(), main(), AtomProbe::marchingCubes(), and AtomProbe::Voxels< T >::setEntry().
◆ getAxisBounds()
| void AtomProbe::Voxels< T >::getAxisBounds | ( | size_t | axis, |
| float & | minV, | ||
| float & | maxV | ||
| ) | const |
Definition at line 828 of file voxels.h.
Referenced by AtomProbe::Voxels< T >::setEntry().
◆ getBinVolume()
| float AtomProbe::Voxels< T >::getBinVolume | ( | ) | const |
Definition at line 1485 of file voxels.h.
References AtomProbe::Voxels< T >::size().
Referenced by AtomProbe::Voxels< T >::clear().
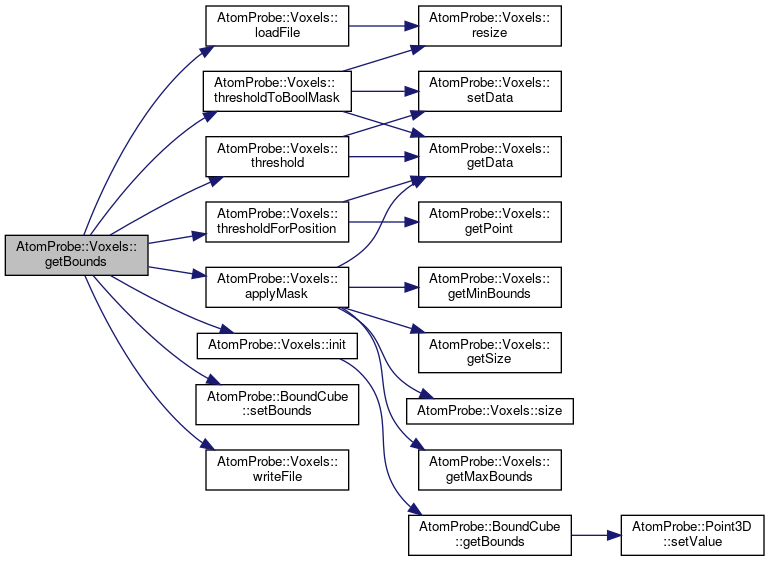
◆ getBounds() [1/2]
|
inline |
Get the bounding size.
Definition at line 252 of file voxels.h.
Referenced by main(), and AtomProbe::marchingCubes().
◆ getBounds() [2/2]
|
inline |
Definition at line 253 of file voxels.h.
References AtomProbe::Voxels< T >::applyMask(), AtomProbe::Voxels< T >::init(), AtomProbe::Voxels< T >::loadFile(), AtomProbe::BoundCube::setBounds(), AtomProbe::Voxels< T >::threshold(), AtomProbe::Voxels< T >::thresholdForPosition(), AtomProbe::Voxels< T >::thresholdToBoolMask(), and AtomProbe::Voxels< T >::writeFile().
◆ getCellUniqueEdgeIndex()
| size_t AtomProbe::Voxels< T >::getCellUniqueEdgeIndex | ( | size_t | x, |
| size_t | y, | ||
| size_t | z, | ||
| unsigned int | edge | ||
| ) | const |
Get a unique integer that corresponds to an edge index for the voxel; where edges are shared between voxels.
Each voxel has 12 edges. These are shared (except voxels that on zero or positive boundary). Return a NON-unique index that corresponds to a specified edge (0->11) Index can be inverted to yield cell
Definition at line 701 of file voxels.h.
Referenced by AtomProbe::Voxels< T >::setEntry().
◆ getData() [1/2]
|
inline |
Retrieve the value of a specific voxel.
Definition at line 1186 of file voxels.h.
References ASSERT.
Referenced by AtomProbe::Voxels< T >::applyMask(), AtomProbe::Voxels< T >::binarise(), AtomProbe::Voxels< T >::countIons(), AtomProbe::Voxels< T >::countPoints(), AtomProbe::Voxels< T >::getInterpolatedData(), AtomProbe::Voxels< T >::getSlice(), AtomProbe::marchingCubes(), AtomProbe::Voxels< T >::resizeKeepData(), AtomProbe::sumVoxels(), AtomProbe::Voxels< T >::threshold(), AtomProbe::Voxels< T >::thresholdForPosition(), and AtomProbe::Voxels< T >::thresholdToBoolMask().
◆ getData() [2/2]
|
inline |
◆ getEdgeCell()
| void AtomProbe::Voxels< T >::getEdgeCell | ( | size_t | edgeUniqId, |
| size_t & | x, | ||
| size_t & | y, | ||
| size_t & | z, | ||
| size_t & | axis | ||
| ) | const |
Convert edge index (only as generted by getCellUniqueEdgeIndex) into a cell & axis value.
Definition at line 751 of file voxels.h.
References ASSERT.
Referenced by AtomProbe::Voxels< T >::getEdgeEnds(), and AtomProbe::Voxels< T >::setEntry().
◆ getEdgeEndApproxVals()
| void AtomProbe::Voxels< T >::getEdgeEndApproxVals | ( | size_t | edgeUniqId, |
| T & | a, | ||
| T & | b | ||
| ) | const |
Return the values that are associated with the edge ends, as returned by getEdgeEnds.
Definition at line 813 of file voxels.h.
References AtomProbe::Voxels< T >::getEdgeEnds(), AtomProbe::Voxels< T >::getIndex(), and AtomProbe::Voxels< T >::getPointData().
Referenced by AtomProbe::marchingCubes(), and AtomProbe::Voxels< T >::setEntry().
◆ getEdgeEnds()
| void AtomProbe::Voxels< T >::getEdgeEnds | ( | size_t | edgeIndex, |
| AtomProbe::Point3D & | a, | ||
| AtomProbe::Point3D & | b | ||
| ) | const |
Convert the edge index (as generated by getEdgeUniqueIndex) into a cenre position.
Definition at line 771 of file voxels.h.
References ASSERT, AtomProbe::BoundCube::containsPt(), AtomProbe::BoundCube::expand(), AtomProbe::Voxels< T >::getEdgeCell(), AtomProbe::Voxels< T >::getMaxBounds(), AtomProbe::Voxels< T >::getMinBounds(), AtomProbe::Voxels< T >::getPitch(), AtomProbe::Voxels< T >::getPoint(), and AtomProbe::BoundCube::setBounds().
Referenced by AtomProbe::Voxels< T >::getEdgeEndApproxVals(), AtomProbe::marchingCubes(), and AtomProbe::Voxels< T >::setEntry().
◆ getIndex()
| void AtomProbe::Voxels< T >::getIndex | ( | size_t & | x, |
| size_t & | y, | ||
| size_t & | z, | ||
| const AtomProbe::Point3D & | p | ||
| ) | const |
Retrieve the XYZ voxel location associated with a given position.
Definition at line 1496 of file voxels.h.
References ASSERT.
Referenced by AtomProbe::Voxels< T >::countIons(), AtomProbe::Voxels< T >::countPoints(), AtomProbe::Voxels< T >::getEdgeEndApproxVals(), AtomProbe::Voxels< T >::getIndexWithUpper(), and AtomProbe::Voxels< T >::getInterpolatedData().
◆ getIndexWithUpper()
| void AtomProbe::Voxels< T >::getIndexWithUpper | ( | size_t & | x, |
| size_t & | y, | ||
| size_t & | z, | ||
| const AtomProbe::Point3D & | p | ||
| ) | const |
Retrieve the XYZ voxel location associated with a given position,.
Definition at line 1515 of file voxels.h.
References AtomProbe::Voxels< T >::getIndex().
◆ getInterpolatedData()
| void AtomProbe::Voxels< T >::getInterpolatedData | ( | const AtomProbe::Point3D & | pt, |
| T & | v | ||
| ) | const |
Definition at line 1678 of file voxels.h.
References ASSERT, AtomProbe::BoundCube::containsPt(), AtomProbe::Voxels< T >::getData(), AtomProbe::Voxels< T >::getIndex(), and AtomProbe::Voxels< T >::getPitch().
Referenced by AtomProbe::Voxels< T >::setEntry().
◆ getInterpSlice()
| void AtomProbe::Voxels< T >::getInterpSlice | ( | size_t | normal, |
| float | offset, | ||
| T * | p, | ||
| size_t | interpMode = VOX_INTERP_NONE |
||
| ) | const |
Definition at line 1614 of file voxels.h.
References ASSERT, AtomProbe::Voxels< T >::getSlice(), AtomProbe::VOX_INTERP_LINEAR, and AtomProbe::VOX_INTERP_NONE.
Referenced by AtomProbe::Voxels< T >::setEntry().

◆ getMaxBounds()
| AtomProbe::Point3D AtomProbe::Voxels< T >::getMaxBounds | ( | ) | const |
Definition at line 954 of file voxels.h.
References ASSERT.
Referenced by AtomProbe::Voxels< T >::applyMask(), AtomProbe::Voxels< T >::getEdgeEnds(), AtomProbe::marchingCubes(), and AtomProbe::Voxels< T >::setEntry().
◆ getMinBounds()
| AtomProbe::Point3D AtomProbe::Voxels< T >::getMinBounds | ( | ) | const |
Get the bounding box vertex (min/max)
Definition at line 947 of file voxels.h.
References ASSERT.
Referenced by AtomProbe::Voxels< T >::applyMask(), AtomProbe::Voxels< T >::getEdgeEnds(), AtomProbe::marchingCubes(), and AtomProbe::Voxels< T >::setEntry().
◆ getPitch()
| AtomProbe::Point3D AtomProbe::Voxels< T >::getPitch | ( | ) | const |
! Get the spacing for a unit cell
Definition at line 480 of file voxels.h.
Referenced by AtomProbe::Voxels< T >::getEdgeEnds(), AtomProbe::Voxels< T >::getInterpolatedData(), AtomProbe::marchingCubes(), and AtomProbe::Voxels< T >::setEntry().
◆ getPoint()
| AtomProbe::Point3D AtomProbe::Voxels< T >::getPoint | ( | size_t | x, |
| size_t | y, | ||
| size_t | z | ||
| ) | const |
Get the position associated with an XYZ voxel.
Definition at line 470 of file voxels.h.
Referenced by AtomProbe::Voxels< T >::getEdgeEnds(), main(), and AtomProbe::Voxels< T >::thresholdForPosition().
◆ getPointData()
| T AtomProbe::Voxels< T >::getPointData | ( | const AtomProbe::Point3D & | pt | ) | const |
Retrieve the value of a datapoint, this is rounded to the nearest voxel.
Definition at line 454 of file voxels.h.
References ASSERT.
Referenced by AtomProbe::Voxels< T >::getEdgeEndApproxVals().
◆ getSize()
| void AtomProbe::Voxels< T >::getSize | ( | size_t & | x, |
| size_t & | y, | ||
| size_t & | z | ||
| ) | const |
Get the size of the data field.
Definition at line 488 of file voxels.h.
References ASSERT.
Referenced by AtomProbe::Voxels< T >::applyMask(), AtomProbe::marchingCubes(), AtomProbe::Voxels< T >::setEntry(), and AtomProbe::sumVoxels().
◆ getSlice()
| void AtomProbe::Voxels< T >::getSlice | ( | size_t | normal, |
| size_t | offset, | ||
| T * | p | ||
| ) | const |
Definition at line 1545 of file voxels.h.
References ASSERT, and AtomProbe::Voxels< T >::getData().
Referenced by AtomProbe::Voxels< T >::getInterpSlice(), and AtomProbe::Voxels< T >::setEntry().
◆ getSum()
| T AtomProbe::Voxels< T >::getSum | ( | const T & | initialVal = T(0.0) | ) | const |
Get the total value of the data field.
An "initial value" is provided to provide the definition of a zero value
Definition at line 1117 of file voxels.h.
References ASSERT.
Referenced by AtomProbe::Voxels< T >::setEntry().
◆ init() [1/2]
| size_t AtomProbe::Voxels< T >::init | ( | size_t | nX, |
| size_t | nY, | ||
| size_t | nZ, | ||
| const BoundCube & | bound | ||
| ) |
Initialise the voxel storage.
Definition at line 982 of file voxels.h.
References AtomProbe::BoundCube::getBounds().
Referenced by AtomProbe::Voxels< T >::getBounds(), and AtomProbe::Voxels< T >::init().

◆ init() [2/2]
| size_t AtomProbe::Voxels< T >::init | ( | size_t | nX, |
| size_t | nY, | ||
| size_t | nZ | ||
| ) |
Initialise the voxel storage.
Definition at line 997 of file voxels.h.
References AtomProbe::Voxels< T >::init().

◆ loadFile()
| size_t AtomProbe::Voxels< T >::loadFile | ( | const char * | cpFilename, |
| size_t | nX, | ||
| size_t | nY, | ||
| size_t | nZ, | ||
| bool | silent = false |
||
| ) |
Load the voxels from file.
Format is flat size_ts in column major return codes: 1: File open error 2: Data size error
Definition at line 1006 of file voxels.h.
References AtomProbe::Voxels< T >::resize(), AtomProbe::VOXELS_BAD_FILE_OPEN, AtomProbe::VOXELS_BAD_FILE_READ, and AtomProbe::VOXELS_BAD_FILE_SIZE.
Referenced by AtomProbe::Voxels< T >::getBounds().
◆ max()
| T AtomProbe::Voxels< T >::max | ( | ) | const |
Find maximum in dataset.
Definition at line 1364 of file voxels.h.
References ASSERT.
Referenced by AtomProbe::Voxels< T >::clear().
◆ min()
| T AtomProbe::Voxels< T >::min | ( | ) | const |
Find minimum in dataset.
Definition at line 1357 of file voxels.h.
References ASSERT.
Referenced by AtomProbe::Voxels< T >::clear().
◆ minMax()
| void AtomProbe::Voxels< T >::minMax | ( | T & | min, |
| T & | max | ||
| ) | const |
Find both min and max in dataset in the same loop.
Definition at line 1372 of file voxels.h.
References ASSERT.
Referenced by AtomProbe::Voxels< T >::clear().
◆ operator/=() [1/2]
| void AtomProbe::Voxels< T >::operator/= | ( | const Voxels< T > & | v | ) |
Element wise division.
Definition at line 1736 of file voxels.h.
References ASSERT.
Referenced by AtomProbe::Voxels< T >::clear().
◆ operator/=() [2/2]
| void AtomProbe::Voxels< T >::operator/= | ( | const T & | v | ) |
◆ operator==()
| bool AtomProbe::Voxels< T >::operator== | ( | const Voxels< T > & | v | ) | const |
Definition at line 1773 of file voxels.h.
Referenced by AtomProbe::Voxels< T >::clear().
◆ resize() [1/2]
| size_t AtomProbe::Voxels< T >::resize | ( | size_t | newX, |
| size_t | newY, | ||
| size_t | newZ, | ||
| const AtomProbe::Point3D & | newMinBound = AtomProbe::Point3D(0.0f,0.0f,0.0f), |
||
| const AtomProbe::Point3D & | newMaxBound = AtomProbe::Point3D(1.0f,1.0f,1.0f) |
||
| ) |
Resize the data field.
This will destroy any data that was already in place If the data needs to be preserved use "resizeKeepData" Data will not be initialised
Definition at line 835 of file voxels.h.
Referenced by AtomProbe::Voxels< T >::binarise(), AtomProbe::Voxels< T >::loadFile(), main(), AtomProbe::marchingCubes(), AtomProbe::Voxels< T >::resize(), AtomProbe::Voxels< T >::resizeKeepData(), AtomProbe::Voxels< T >::setEntry(), and AtomProbe::Voxels< T >::thresholdToBoolMask().
◆ resize() [2/2]
| size_t AtomProbe::Voxels< T >::resize | ( | const Voxels< T > & | v | ) |
Definition at line 858 of file voxels.h.
References AtomProbe::Voxels< T >::resize().
◆ resizeKeepData()
| size_t AtomProbe::Voxels< T >::resizeKeepData | ( | size_t | newX, |
| size_t | newY, | ||
| size_t | newZ, | ||
| unsigned int | direction = CLIP_LOWER_SOUTH_WEST, |
||
| const AtomProbe::Point3D & | newMinBound = AtomProbe::Point3D(0.0f,0.0f,0.0f), |
||
| const AtomProbe::Point3D & | newMaxBound = AtomProbe::Point3D(1.0f,1.0f,1.0f), |
||
| const T & | fill = T(0), |
||
| bool | doFill = false |
||
| ) |
Resize the data field, maintaining data as best as possible.
This will preserve data by resizing as much as possible about the origin. If the bounds are extended, the "fill" value is used by default iff doFill is set to true.
Definition at line 864 of file voxels.h.
References ASSERT, AtomProbe::CLIP_LOWER_SOUTH_WEST, AtomProbe::Voxels< T >::getData(), AtomProbe::Voxels< T >::resize(), AtomProbe::Voxels< T >::setData(), and AtomProbe::Voxels< T >::swap().
Referenced by AtomProbe::Voxels< T >::setEntry().
◆ setBounds() [1/2]
| void AtomProbe::Voxels< T >::setBounds | ( | const AtomProbe::Point3D & | pMin, |
| const AtomProbe::Point3D & | pMax | ||
| ) |
Set the bounding size.
Definition at line 961 of file voxels.h.
References ASSERT.
Referenced by main(), AtomProbe::Voxels< T >::setBounds(), and AtomProbe::Voxels< T >::setEntry().
◆ setBounds() [2/2]
| void AtomProbe::Voxels< T >::setBounds | ( | const BoundCube & | b | ) |
Definition at line 976 of file voxels.h.
References AtomProbe::BoundCube::max(), AtomProbe::BoundCube::min(), and AtomProbe::Voxels< T >::setBounds().
◆ setData() [1/2]
| void AtomProbe::Voxels< T >::setData | ( | size_t | x, |
| size_t | y, | ||
| size_t | z, | ||
| const T & | val | ||
| ) |
Retrieve a reference to the data ata given position.
Set the value of a point in the dataset
Definition at line 435 of file voxels.h.
References ASSERT.
Referenced by AtomProbe::Voxels< T >::binarise(), AtomProbe::Voxels< T >::countIons(), AtomProbe::Voxels< T >::countPoints(), main(), AtomProbe::marchingCubes(), AtomProbe::Voxels< T >::resizeKeepData(), AtomProbe::Voxels< T >::setEntry(), AtomProbe::Voxels< T >::threshold(), and AtomProbe::Voxels< T >::thresholdToBoolMask().
◆ setData() [2/2]
|
inline |
◆ setEntry()
|
inline |
Definition at line 158 of file voxels.h.
References AtomProbe::CLIP_LOWER_SOUTH_WEST, AtomProbe::Voxels< T >::count(), AtomProbe::Voxels< T >::deprecatedGetEdgeUniqueIndex(), AtomProbe::Voxels< T >::fill(), AtomProbe::Voxels< T >::getAxisBounds(), AtomProbe::Voxels< T >::getCellUniqueEdgeIndex(), AtomProbe::Voxels< T >::getEdgeCell(), AtomProbe::Voxels< T >::getEdgeEndApproxVals(), AtomProbe::Voxels< T >::getEdgeEnds(), AtomProbe::Voxels< T >::getInterpolatedData(), AtomProbe::Voxels< T >::getInterpSlice(), AtomProbe::Voxels< T >::getMaxBounds(), AtomProbe::Voxels< T >::getMinBounds(), AtomProbe::Voxels< T >::getPitch(), AtomProbe::Voxels< T >::getSize(), AtomProbe::Voxels< T >::getSlice(), AtomProbe::Voxels< T >::getSum(), AtomProbe::Voxels< T >::resize(), AtomProbe::Voxels< T >::resizeKeepData(), AtomProbe::Voxels< T >::setBounds(), AtomProbe::Voxels< T >::setData(), and AtomProbe::VOX_INTERP_NONE.
◆ setPoint()
| void AtomProbe::Voxels< T >::setPoint | ( | const AtomProbe::Point3D & | pt, |
| const T & | val | ||
| ) |
◆ size()
|
inline |
Definition at line 361 of file voxels.h.
Referenced by AtomProbe::Voxels< T >::applyMask(), AtomProbe::Voxels< T >::calculateDensity(), AtomProbe::Voxels< T >::getBinVolume(), and AtomProbe::sumVoxels().
◆ sizeofType()
|
inlinestatic |
Return the sizeof value for the T type.
Maybe there is a better way to do this, I don't know
Definition at line 300 of file voxels.h.
References AtomProbe::Voxels< T >::binarise().
◆ swap()
| void AtomProbe::Voxels< T >::swap | ( | Voxels< T > & | v | ) |
Swap object contents with other voxel object.
Definition at line 1173 of file voxels.h.
Referenced by AtomProbe::Voxels< T >::resizeKeepData().
◆ threshold()
| void AtomProbe::Voxels< T >::threshold | ( | const T & | thresh, |
| bool | keepUpper, | ||
| const T & | newVal | ||
| ) |
Threshold the voxels, keeping either the lower or upper values above this threshold.
Definition at line 1238 of file voxels.h.
References AtomProbe::Voxels< T >::getData(), and AtomProbe::Voxels< T >::setData().
Referenced by AtomProbe::Voxels< T >::getBounds().
◆ thresholdForPosition()
| void AtomProbe::Voxels< T >::thresholdForPosition | ( | std::vector< AtomProbe::Point3D > & | p, |
| const T & | thresh, | ||
| bool | lowerEq = false |
||
| ) | const |
Find the positions of the voxels that are above or below a given value.
Returns the positions of the voxels' centroids for voxels that have, by default, a value greater than that of thresh. This behaviour can by reversed to "lesser than" by setting lowerEq
Definition at line 1193 of file voxels.h.
References AtomProbe::Voxels< T >::getData(), and AtomProbe::Voxels< T >::getPoint().
Referenced by AtomProbe::Voxels< T >::getBounds().
◆ thresholdToBoolMask()
| void AtomProbe::Voxels< T >::thresholdToBoolMask | ( | const T & | thresh, |
| bool | keepUpper, | ||
| Voxels< bool > & | result | ||
| ) | const |
Generate a boolean voxel field stating whether the data is above or below the threshold specified.
Definition at line 1274 of file voxels.h.
References AtomProbe::Voxels< T >::getData(), AtomProbe::Voxels< T >::resize(), AtomProbe::Voxels< T >::setData(), and XOR.
Referenced by AtomProbe::Voxels< T >::getBounds().
◆ trapezIntegral()
| T AtomProbe::Voxels< T >::trapezIntegral | ( | ) | const |
Integrate the datataset via the trapezoidal method.
Definition at line 1130 of file voxels.h.
Referenced by AtomProbe::Voxels< T >::clear().
◆ writeFile()
| size_t AtomProbe::Voxels< T >::writeFile | ( | const char * | cpFilename | ) | const |
Write the voxel objects in column major written out to file.
Format is flat objects ("T"s) in column major format. Returns nonzero on failure
Definition at line 1093 of file voxels.h.
References ASSERT.
Referenced by AtomProbe::Voxels< T >::getBounds().
The documentation for this class was generated from the following file:
- include/atomprobe/helper/voxels.h
 1.8.13
1.8.13