Classes | |
| class | AbundanceData |
| Class to load abundance information for natural isotopes. More... | |
| struct | ATO_ENTRY |
| class | AxisCompare |
| Functor allowing for sorting of points in 3D. More... | |
| struct | BACKGROUND_PARAMS |
| class | BodyCentredCubicGen |
| class | BoundCube |
| Helper class to define a bounding cube. More... | |
| class | ComparePairFirst |
| class | ComparePairFirstReverse |
| class | ComparePairSecond |
| class | CrystalGen |
| struct | DATA_FIT |
| class | EPOS_ENTRY |
| class | FaceCentredCubicGen |
| struct | FLATTENED_RANGE |
| Structure that allows for the multirange data to be mapped into. More... | |
| class | GnomonicProjection |
| class | IonHit |
| This is a data holding class for POS file ions, from. More... | |
| struct | IONHIT |
| Record as stored in a .POS file. More... | |
| struct | ISOTOPE_ENTRY |
| class | K3DNodeApprox |
| Node Class for storing point. More... | |
| class | K3DNodeExact |
| Node Class for storing point. More... | |
| class | K3DTreeApprox |
| 3D specific KD tree More... | |
| class | K3DTreeExact |
| 3D specific KD tree More... | |
| class | LibVersion |
| Class to hold the library version data. More... | |
| class | LINE |
| class | LinearFeedbackShiftReg |
| This class implements a Linear Feedback Shift Register (in software) More... | |
| class | MassTool |
| Class that brute-force solves the knapsack problem. More... | |
| class | Mesh |
| Simple mesh class for storing and manipulating meshes consisting of 2 and 3D simplexes (triangles or tetrahedra) More... | |
| class | MILLER_TRIPLET |
| class | ModifiedFocusSphericProjection |
| class | MultiRange |
| Data storage and retrieval class for "ranging" a spectra, where overlapping ranges are permitted. More... | |
| struct | OVERLAP_PROBLEM_SETTINGS |
| class | Point3D |
| A 3D point data class storage. More... | |
| struct | Point3f |
| Data storage structure for points. More... | |
| class | ProgressBar |
| struct | Quaternion |
| Data storage structure for quaternions. More... | |
| class | RandNumGen |
| struct | RANGE_MOLECULE |
| class | RangeFile |
| Data storage and retrieval class for various range files. More... | |
| class | ReconstructionSphereOnCone |
| class | RGBf |
| Data holder for colour as float. More... | |
| struct | SIMPLE_SPECIES |
| class | SimpleCubicGen |
| struct | SINGLE_HIT |
| Single 3DAP hit event. More... | |
| class | SphericPlaneProjection |
| struct | SphericProjectionParams |
| class | StereographicProjection |
| class | TETRAHEDRON |
| struct | THREEDAP_DATA |
| experimental setup data More... | |
| struct | THREEDAP_EXPERIMENT |
| Data structure that contains the experiment information present in a 3Dap file. More... | |
| class | TRIANGLE |
| struct | TriangleWithIndexedVertices |
| class | TriangleWithVertexNorm |
| struct | VOLTAGE_DATA |
| Voltage data structure – these updates occur periodically during the experiment. More... | |
| class | Voxels |
| Template class that stores 3D voxel data. More... | |
| class | Weight |
| Placeholder class for containing input weights for MassTool. More... | |
| struct | ZECH_ROOT |
Typedefs | |
| typedef struct AtomProbe::ATO_ENTRY | ATO_ENTRY |
| typedef struct AtomProbe::IONHIT | IONHIT |
| Record as stored in a .POS file. More... | |
Functions | |
| unsigned int | generate1DAxialDistHist (const std::vector< Point3D > &pointList, K3DTreeExact &tree, const Point3D &axisDir, float distMax, std::vector< unsigned int > &histogram) |
| Generate a 1D axial distribution function,. More... | |
| unsigned int | generate1DAxialDistHistSweep (const std::vector< Point3D > &pointList, K3DTreeExact &tree, float distMax, float dTheta, float dPhi, AtomProbe::ProgressBar &prog, std::vector< std::vector< std::vector< unsigned int > > > &histogram) |
| Generate a series of 1D distribution functions, one per pixel in a 2D grid of spherical coordinate directions. More... | |
| void | computeIonDistAdjacency (const MultiRange &mrf, const AbundanceData &abundance, const OVERLAP_PROBLEM_SETTINGS &settings, gsl_matrix *&m, gsl_vector *&vOwnership, gsl_vector *&vMass) |
| Calculate the adjacency matrix for the ions in a multirange. More... | |
| unsigned int | GetReducedHullPts (const std::vector< IonHit > &points, float reductionDim, unsigned int *progress, bool(*callback)(bool), std::vector< IonHit > &pointResult) |
| Obtain a set of points that are a subset of points the convex hull that are at least "reductionDim" away from the hull itself. More... | |
| unsigned int | computeConvexHull (const std::vector< IonHit > &data, unsigned int *progress, bool(*callback)(bool), std::vector< Point3D > &curHull, bool freeHull) |
| Obtain the convex hull of a set of ions. More... | |
| void | filterPeakNeedBiggerObs (const AtomProbe::AbundanceData &massTable, const std::vector< float > &peakData, float tolerance, size_t solutionCharge, std::vector< std::vector< AtomProbe::ISOTOPE_ENTRY > > &solutions) |
| void | filterBySolutionPPM (const AtomProbe::AbundanceData &massTable, float minPpm, std::vector< std::vector< AtomProbe::ISOTOPE_ENTRY > > &solutions) |
| Use the maximum possible PPM for each isotopic combination to filter possible solutions. More... | |
| std::vector< float > | maxExplainedFraction (const std::vector< std::pair< float, float > > &massData, float peakMass, float massWidth, const std::vector< std::vector< AtomProbe::ISOTOPE_ENTRY > > &solutions, const AtomProbe::AbundanceData &massTable, float massDistTol, unsigned int solutionCharge) |
| Compute the fraction of the data that has been explained, using the natural abundance information, and intensity data. More... | |
| bool | correlationHistogram (const std::vector< EPOS_ENTRY > &eposIons, std::vector< std::vector< unsigned int > > &histogram, float stepMass, float endMass, bool lowerTriangular=false) |
| Generates an ion "correlation histogram" from a vector of EPOS ions. The input vector MUST More... | |
| bool | accumulateCorrelationHistogram (const std::vector< EPOS_ENTRY > &eposIons, std::vector< std::vector< unsigned int > > &histogram, float stepMass, float endMass, bool lowerTriangular=true) |
| Increments a correlation histogram, as per correlationHistogram,. More... | |
| template<class T > | |
| void | linearHistogram (const std::vector< T > &data, T start, T end, T step, std::vector< T > &histVals) |
| void | marchingCubes (const Voxels< float > &v, float isoValue, std::vector< TriangleWithVertexNorm > &tVec) |
| void | checkMassRangingCorrectness (const AtomProbe::RangeFile &rng, AtomProbe::AbundanceData &massTable, float massTolerance, unsigned int maxChargeState, unsigned int maxComponents, std::vector< bool > &badRanges) |
| Ensure that each mass given spans a peak that should exist. More... | |
| void | computeRotationMatrix (const Point3D &ur1, const Point3D &ur2, const Point3D &r1, const Point3D &r2, gsl_matrix *m) |
| void | computeRotationMatrix (const Point3D &ur1, const Point3D &ur2, const Point3D &r1, const Point3D &r2, std::vector< std::vector< float > > &m) |
| Convenience wrapper for conputeRotationMatrix, which uses vector containers. More... | |
| unsigned int | computeRotationMatrixWahba (const std::vector< Point3D > &unrotated, const std::vector< Point3D > &rotated, const std::vector< float > &weights, gsl_matrix *&R) |
| Given a series of paired observations of directions, compute the least squares rotation matrix between them. More... | |
| bool | leastSquaresOverlapSolve (const AtomProbe::MultiRange &rangeData, const AtomProbe::AbundanceData &abundance, const std::vector< IonHit > &hits, float(*backgroundEstimator)(float, float), std::vector< std::pair< unsigned int, float > > &decomposedHits, float rangeTolerance=0) |
| Solve overlaps using least squares for given multi-range. More... | |
| void | setHardAssert (bool enabled) |
| Do assertions cause a straight up crash (enabled), or write a mesage to stderr (disabled)? By default, hard crash. More... | |
| void | askAssert (const char *, unsigned int) |
| Either abort program, or ask the user what to do for an assertion. depending on hardAssert setting. More... | |
| void | computeComposition (const std::vector< unsigned int > &countData, std::vector< float > &compositionData) |
| void | convertEPOStoPos (const std::vector< EPOS_ENTRY > &eposEntry, std::vector< IonHit > &posFile) |
| Convert an incoming entry of EPOS files to pos, and the append this to the pos vector given. More... | |
| void | gsl_matrix_mult (const gsl_matrix *A, const gsl_matrix *B, gsl_matrix *&res, bool alloc=false) |
| unsigned int | estimateRank (const gsl_matrix *m, float tolerance=sqrt(std::numeric_limits< float >::epsilon())) |
| Estimate the rank of the given matrix. More... | |
| bool | solveLeastSquares (gsl_matrix *m, gsl_vector *b, gsl_vector *&x) |
| Use an SVD based least-squares solver to solve Mx=b (for x). More... | |
| template<class T > | |
| T | weightedMean (const std::vector< T > &values, const std::vector< T > &weight) |
| template<typename T > | |
| void | meanAndStdev (const std::vector< T > &f, float &meanVal, float &stdevVal, bool normalCorrection=true) |
| unsigned int | vectorPointDir (const Point3D &pA, const Point3D &pB, const Point3D &vC, const Point3D &vD) |
| Check which way vectors attached to two 3D points "point",. More... | |
| float | distanceToSegment (const Point3D &fA, const Point3D &fB, const Point3D &p) |
| float | signedDistanceToFacet (const Point3D &fA, const Point3D &fB, const Point3D &fC, const Point3D &normal, const Point3D &p) |
| Find the distance between a point, and a triangular facet – may be positive or negative. More... | |
| float | distanceToFacet (const Point3D &fA, const Point3D &fB, const Point3D &fC, const Point3D &normal, const Point3D &p) |
| float | dotProduct (float a1, float a2, float a3, float b1, float b2, float b3) |
| Inline func for calculating a(dot)b. More... | |
| double | det3by3 (const double *ptArray) |
| double | pyramidVol (const Point3D *planarPts, const Point3D &apex) |
| bool | triIsDegenerate (const Point3D &fA, const Point3D &fB, const Point3D &fC) |
| void | quat_rot (Point3D &p, const Point3D &r, float angle) |
| Rotate a point around a given rotation axis by a specified angle. More... | |
| void | quat_rot (Point3f *point, const Point3f *rotVec, float angle) |
| Rotate a point around a given vector, with specified angle. More... | |
| void | quat_rot_array (Point3f *point, unsigned int n, const Point3f *rotVec, float angle) |
| Rotate each point in array of size n around a given vector, with specified angle. More... | |
| void | quat_rot_array (Point3D *point, unsigned int n, const Point3f *rotVec, float angle) |
| Rotate each point in array of size n around a given vector, with specified angle. More... | |
| void | quat_get_rot_quat (const Point3f *rotVec, float angle, Quaternion *rotQuat) |
| Compute the quaternion for specified rotation. More... | |
| void | quat_rot_apply_quat (Point3f *point, const Quaternion *rotQuat) |
| Use previously generated quats from quat_get_rot_quats to rotate a point. More... | |
| template<class T > | |
| bool | tolEqual (const T &a, const T &b, const T &f) |
| Test for equality within tolerance f (||a-b|| < f) More... | |
| template<class T > | |
| void | selectElements (const std::vector< T > &in, const std::vector< unsigned int > &indices, std::vector< T > &out) |
| Obtain the elements at positions indicies in the input vector, copy to output. More... | |
| template<class T > | |
| void | transposeVector (std::vector< std::vector< T > > &v) |
| Perform an out-of-place tranposition of a given vector. More... | |
| template<class T > | |
| void | vectorMultiErase (std::vector< T > &vec, const std::vector< bool > &wantKill) |
| Remove elements from the vector, without preserving order. More... | |
| void | sampleIons (const std::vector< IonHit > &ions, float sampleFactor, std::vector< IonHit > &sampled, bool strongRandom=true) |
| template<class T > | |
| size_t | randomSelect (std::vector< T > &result, const std::vector< T > &source, size_t num, gsl_rng *rng) |
| template<class T > | |
| void | randomIndices (std::vector< T > &res, size_t num, size_t nMax, gsl_rng *rng) |
| template<class T1 , class T2 > | |
| bool | stream_cast (T1 &result, const T2 &obj) |
| Template function to cast and object to another by the stringstream. More... | |
| template<class T > | |
| void | strAppend (std::string s, const T &a) |
| void | genColString (unsigned char r, unsigned char g, unsigned char b, unsigned char a, std::string &s) |
| void | genColString (unsigned char r, unsigned char g, unsigned char b, std::string &s) |
| bool | parseColString (const std::string &str, unsigned char &r, unsigned char &g, unsigned char &b, unsigned char &a) |
| Parse a colour string containing rgb[a]; hex for with leading #. More... | |
| std::string | digitString (unsigned int thisDigit, unsigned int maxDigit) |
| Generate a string with leading digits up to maxDigit (eg, if maxDigit is 424, and thisDigit is 1. More... | |
| std::string | stripWhite (const std::string &str) |
| Strip whitespace, (eg tab,space) from either side of a string. More... | |
| std::string | stripChars (const std::string &Str, const char *chars) |
| std::string | lowercase (std::string s) |
| Return a lowercase version for a given string. More... | |
| std::string | uppercase (std::string s) |
| Return a uppercase version for a given string. More... | |
| void | stripZeroEntries (std::vector< std::string > &s) |
| void | splitStrsRef (const char *cpStr, const char delim, std::vector< std::string > &v) |
| Split string references using a single delimiter. More... | |
| void | splitStrsRef (const char *cpStr, const char *delim, std::vector< std::string > &v) |
| Split string references using any of a given string of delimiters. More... | |
| std::string | onlyFilename (const std::string &path) |
| Return only the filename component. More... | |
| std::string | onlyDir (const std::string &path) |
| Return only the directory name component of the full path. More... | |
| std::string | convertFileStringToNative (const std::string &s) |
| Convert a path format into a native path from unix format. More... | |
| std::string | convertFileStringToCanonical (const std::string &s) |
| Convert a path format into a unix path from native format. More... | |
| std::string | tabs (unsigned int nTabs) |
| std::string | stlWStrToStlStr (const std::wstring &s) |
| std::wstring | stlStrToStlWStr (const std::string &s) |
| void | nullifyMarker (char *buffer, char marker) |
| template<class T , class U > | |
| void | castVoxels (const Voxels< T > &src, Voxels< U > &dest) |
| Convert one type of voxel into another by assignment operator. More... | |
| template<class T , class U > | |
| void | sumVoxels (const Voxels< T > &src, U &counter) |
| Use one counting type to sum counts in a voxel of given type. More... | |
| unsigned int | XMLHelpNextType (xmlNodePtr &node, int) |
| unsigned int | XMLHelpFwdToElem (xmlNodePtr &node, const char *nodeName) |
| unsigned int | XMLHelpFwdNotElem (xmlNodePtr &node, const char *nodeName) |
| unsigned int | XMLHelpFwdToList (xmlNodePtr &node, const std::vector< std::string > &nodeList) |
| std::string | XMLHelpGetText (xmlNodePtr &node) |
| void | XMLFreeDoc (void *data) |
| Free a xmlDoc pointer. For use in conjunction with std::unique_ptr for auto-deallocation. More... | |
| template<class T > | |
| unsigned int | XMLHelpGetProp (T &prop, xmlNodePtr node, std::string propName) |
| template<class T > | |
| bool | XMLGetNextElemAttrib (xmlNodePtr &nodePtr, T &v, const char *nodeName, const char *attrib) |
| Grab the specified attribute from the next element, then stream_cast() it into the passed-in object. Returns true on success, false on error. More... | |
| const char * | getRecordReadErrString (unsigned int errCode) |
| const char * | getAtoErrString (unsigned int errCode) |
| unsigned int | loadPosFile (std::vector< IonHit > &posIons, const char *posFile) |
| Load a pos file directly into a single ion list. More... | |
| unsigned int | loadPosFile (std::vector< IonHit > &posIons, const char *posFile, unsigned int nSamplesMax) |
| As per loadPosFile, but with an additional setting for the maximum number of ions to load. More... | |
| const char * | getPosFileErrString (unsigned int errMesg) |
| unsigned int | savePosFile (const std::vector< Point3D > &points, float mass, const char *name, bool append=false) |
| Save a vector of Point3Ds into a pos file, using a fixed mass, return nonzero on error. More... | |
| unsigned int | savePosFile (const std::vector< IonHit > &data, const char *name, bool append=false) |
| Save a vector of IonHits into a "pos" file, return nonzero on error. More... | |
| unsigned int | saveTapsimBin (const std::vector< IonHit > &posIons, std::ostream &f) |
| Write a tapsim file from a vector of IonHits. More... | |
| unsigned int | saveTapsimBin (const std::vector< IonHit > &posIons, const char *filename) |
| Write a tapsim file from a vector of ionHits. More... | |
| size_t | loadEposFile (std::vector< EPOS_ENTRY > &outData, const char *filename) |
| Load an entire "EPOS" File. More... | |
| size_t | chunkLoadEposFile (std::vector< EPOS_ENTRY > &outData, const char *filename, unsigned int chunkSize, unsigned int chunkOffset, unsigned int &nEntriesLeft) |
| Load an "EPOS" file, with a maximum chunk size. More... | |
| unsigned int | loadTextData (const char *cpFilename, std::vector< std::vector< float > > &dataVec, std::vector< std::string > &headerVec, const char *delim="\", bool allowNan=true, bool allowConvFails=false, unsigned int headerCount=-1) |
| Load a CSV, TSV or similar text file. Assumes "C" Locale for input (ie "." as decimal separator). More... | |
| unsigned int | readPosapOps (const char *file, THREEDAP_EXPERIMENT &data, unsigned int &badLine, unsigned int &progress, std::atomic< bool > &wantAbort, unsigned int nDelayLines=2, bool strictMode=false) |
| Function to read POSAP "OPS" files. More... | |
| unsigned int | loadATOFile (const char *fileName, std::vector< ATO_ENTRY > &ions, unsigned int forceEndian=0) |
| Load a LAWATAP "ATO" file. More... | |
| bool | operator< (const SIMPLE_SPECIES &a, const SIMPLE_SPECIES &b) |
| unsigned int | parseCompositionData (const std::vector< std::string > &s, const AbundanceData &atomTable, std::map< unsigned int, double > &atomData) |
| Convert atomic string label data, in the form "Fe 0.81" to fraction map. More... | |
| void | convertMolToMass (const AbundanceData &atomTable, const std::map< unsigned int, double > &compositionMols, std::map< unsigned int, double > &compositionMass) |
| Convert the given composition from molar fraction data to mass fraction. More... | |
| void | convertMassToMol (const AbundanceData &atomTable, const std::map< unsigned int, double > &compositionMass, std::map< unsigned int, double > &compositionMols) |
| Convert the given composition from mass fraction data to molar fraction. More... | |
| void | findOverlaps (const AbundanceData &natData, const RangeFile &rng, float massDelta, unsigned int maxCharge, std::vector< std::pair< size_t, size_t > > &overlapIdx, std::vector< std::pair< float, float > > &overlapMasses) |
| Find the overlaps stemming from a given rangefile. More... | |
| void | findOverlaps (const AbundanceData &natData, const std::vector< std::string > &ions, float massDelta, unsigned int maxCharge, std::vector< std::pair< size_t, size_t > > &overlapIdx, std::vector< std::pair< float, float > > &overlapMasses) |
| As per findOverlaps(...Rangefile ...) , but uses a vector of ions instead. More... | |
| void | findOverlapsFromSpectra (const std::vector< std::vector< std::pair< float, float > > > &massDistributions, float massDelta, const std::set< unsigned int > &skipIDs, std::vector< std::pair< size_t, size_t > > &overlapIdx, std::vector< std::pair< float, float > > &overlapMasses) |
| Find overlaps in the given mass distributions, as per findOverlaps(...Rangefile...) More... | |
| bool | reconstructTest () |
| bool | fitVoigt (const std::vector< double > &x, const std::vector< double > &y, double &sigma, double &gamma, double &mu, double &, bool autoInit=true) |
| Fit a Voigt function to the given X/Y values. Internally, a function minimiser is used. More... | |
| void | voigtProfile (const std::vector< double > &x, double sigma, double gamma, double mu, double amp, std::vector< double > &y) |
| Generate a shifted voigt profile. More... | |
| bool | fitDoniachSunjic (const std::vector< double > &x, const std::vector< double > &y, double &a, double &f, double &mu, double &, bool autoInit=true) |
| Fit a Doniach-Sunjic curve. More... | |
| void | doniachSunjic (const std::vector< double > &x, double &a, double &F, double &mu, double &, std::vector< double > &y) |
| generate a Doniach-Sunjic profile More... | |
| bool | fitLikeLogGaussian (const std::vector< double > &x, const std::vector< double > &y, double &lambda, double &sigma, double &xp, double &, double &h, bool autoInit=true) |
| Fit a smoothed log-gaussian curve (arxiv:0711.4449) More... | |
| void | likeLogGaussian (const std::vector< double > &x, double &xp, double &sigma, double &lambda, double &, double &h, std::vector< double > &y) |
| Generate a smoothed log-gaussian curve. More... | |
| bool | fitExpNorm (const std::vector< double > &x, const std::vector< double > &y, double &K, double &mu, double &sigma, double &, bool autoInit=true) |
| Fit a smoothed log-gaussian curve (arxiv:0711.4449) More... | |
| void | expNorm (const std::vector< double > &x, double &K, double &mu, double &sigma, double &, std::vector< double > &y) |
| Exponentially decaying normal distribution. More... | |
| double | lsq (const std::vector< double > &y, const std::vector< double > &yFit) |
| unsigned int | doFitBackground (const std::vector< float > &massData, BACKGROUND_PARAMS ¶ms) |
| Perform a background fit, assuming constant TOF noise, from 0->inf time. More... | |
| std::string | getFitErrorMsg (unsigned int errCode) |
| void | createMassBackground (float massStart, float massEnd, unsigned int nBinsMass, float tofBackIntensity, std::vector< float > &histogram) |
| Build a histogram of the background counts. More... | |
| void | findPeaks (const std::vector< float > &x0, std::vector< unsigned int > &peakInds, float sel, bool autoSel=true, bool includeEndpoints=true) |
| Simple peak-finding algorithm. More... | |
| void | poissonConfidenceObservation (float counts, float alpha, float &lBound, float &uBound) |
| Obtain poisson confidence limits for the mean of a poisson distribution. More... | |
| bool | numericalEstimatePoissRatioConf (float lambda1, float lambda2, float alpha, unsigned int nTrials, gsl_rng *r, float &lBound, float &uBound) |
| Brute-force poisson ratio confidence estimator. Returns the confidence interval in the estimate of the mean, at the given rates. More... | |
| bool | numericalEstimateGaussRatioConf (float mu1, float mu2, float var1, float var2, float alpha, unsigned int nTrials, gsl_rng *r, float &lBound, float &uBound) |
| Brute-force guassian ratio confidence estimator. More... | |
| bool | numericalEstimateSkellamConf (float lambda1, float lambda2, float alpha, unsigned int nTrials, gsl_rng *r, float &lBound, float &uBound) |
| Brute-force the confidence bounds of a skellam distribution. More... | |
| bool | zechConfidenceLimits (float lambdaBack, unsigned int observation, float alpha, float &estimate) |
| Provides a best estimate for true signal when true signal, background. More... | |
| bool | rangeOverlaps (const pair< float, float > &r1, const pair< float, float > &r2) |
| void | computeRangeAdjacency (const MultiRange &mrf, const OVERLAP_PROBLEM_SETTINGS &settings, gsl_matrix *&m) |
| void | freeConvexHull () |
| unsigned int | doHull (unsigned int bufferSize, double *buffer, vector< Point3D > &resHull, Point3D &midPoint, bool freeHullOnExit) |
| unsigned int | countIntensityEvents (const vector< pair< float, float > > &data, float minV, float maxV) |
| void | buildFrequencyTable (const vector< ISOTOPE_ENTRY > &solutionVec, vector< size_t > &solutionElements, vector< size_t > &solutionFrequency) |
| vector< float > | maxExplainedFraction (const vector< pair< float, float > > &intensityData, float peakMass, float massWidth, const vector< vector< ISOTOPE_ENTRY > > &solutions, const AbundanceData &massTable, float massDistTol, unsigned int solutionCharge) |
| bool | incrementCorrelationHist (const std::vector< EPOS_ENTRY > &eposIons, std::vector< std::vector< unsigned int > > &histogram, float stepMass, float endMass) |
| template<class T > | |
| void | removeElements (const std::vector< size_t > &elems, std::vector< T > &vec) |
| bool | pairContains (const pair< float, float > &p, float m) |
| RANGE_MOLECULE | getRangeMolecule (const AbundanceData &massTable, const RangeFile &rng, unsigned int rangeId) |
| double | gsl_matrix_det (gsl_matrix *A, bool inPlace=false) |
| gsl_matrix * | getRotationMatrix (const Point3D &pt, float theta) |
| float | nullBackground (float rangeStart, float rangeEnd) |
| bool | getHardAssert () |
| char * | myStrDup (const char *s) |
| void | pushLocale (const char *newLocale, int type) |
| void | popLocale () |
| bool | getFilesize (const char *fname, size_t &size) |
| void | gsl_print_matrix (const gsl_matrix *m) |
| void | gsl_print_vector (const gsl_vector *v) |
| float | gsl_determinant (const gsl_matrix *m) |
| bool | isNotDirectory (const char *filename) |
| int | fpeek (FILE *stream) |
| void | quat_mult_no_second_a (Quaternion *result, const Quaternion *q1, const Quaternion *q2) |
| void | quat_pointmult (Point3f *result, const Quaternion *q1, const Quaternion *q2) |
| void | quat_invert (Quaternion *quat) |
| void | ucharToHexStr (unsigned char c, std::string &s) |
| void | hexStrToUChar (const std::string &s, unsigned char &c) |
| string | XMLHelpGetText (xmlNodePtr node) |
| template<class T > | |
| unsigned int | fixedRecordReader (const char *filename, bool(*recordReader)(const char *bufRead, const char *destBuf), size_t recordSize, std::vector< T > &outputData) |
| template<class T > | |
| unsigned int | fixedRecordChunkReader (const char *filename, bool(*recordReader)(const char *bufRead, const char *destBuf), size_t recordSize, std::vector< T > &outputData, unsigned int chunkSize, unsigned int chunkOffset, unsigned int &nEntriesLeft) |
| unsigned int | loadTapsimBinFile (vector< IonHit > &posIons, const char *posfile) |
| bool | readEposRecord (const char *src, const char *dest) |
| bool | strhas (const char *cpTest, const char *cpPossible) |
| template<class T > | |
| void | linkIdentifiers (vector< T > &link) |
| bool | pairOverlaps (float aStart, float aEnd, float bStart, float bEnd) |
| bool | isValidElementSymbol (std::string &symbol) |
| string | rangeToStr (const pair< float, float > &rng) |
| bool | matchComposedName (const std::map< string, size_t > &composedNames, const vector< pair< string, size_t > > &namesToFind, size_t &matchOffset) |
| void | findOverlapsFromSpectra (const vector< vector< pair< float, float > > > &massDistributions, float massDelta, const set< unsigned int > &skipIDs, vector< pair< size_t, size_t > > &overlapIdx, vector< pair< float, float > > &overlapMasses) |
| void | findOverlaps (const AbundanceData &natData, const RangeFile &rng, float massDelta, unsigned int maxCharge, vector< pair< size_t, size_t > > &overlapIdx, vector< pair< float, float > > &overlapMasses) |
| void | findOverlaps (const AbundanceData &natData, const vector< string > &ionNames, float massDelta, unsigned int maxCharge, vector< pair< size_t, size_t > > &overlapIdx, vector< pair< float, float > > &overlapMasses) |
| int | gcd (int a, int b) |
| std::ostream & | operator<< (std::ostream &stream, const BoundCube &b) |
| std::ostream & | operator<< (std::ostream &stream, const IonHit &ion) |
| bool | antiRotateMatch (const unsigned int *a, const unsigned int *b, size_t n) |
| bool | rotateMatch (const unsigned int *a, const unsigned int *b, size_t n, bool directionForwards=true) |
| float | signVal (unsigned int val) |
| size_t | findMaxLessThanOrEq (const vector< std::pair< size_t, size_t > > &v, size_t value) |
| float | Determinant (float **a, int n) |
| float | fourDeterminant (const Point3D &a, const Point3D &b, const Point3D &c, const Point3D &d) |
| unsigned int | edgeIdx (unsigned int i, unsigned int j) |
| int | intersect_RayTriangle (const Point3D &rayStart, const Point3D &rayEnd, Point3D *tri, Point3D &I) |
| void | findNearVerticies (float tolerance, const vector< Point3D > &ptVec, vector< std::pair< size_t, vector< size_t > > > &clusterList) |
| void | findNearVerticies (float tolerance, const vector< Point3D > &ptVec, std::list< std::pair< size_t, vector< size_t > > > &clusterList) |
| std::ostream & | operator<< (std::ostream &stream, const Point3D &pt) |
| double | SphericProjectionEqn (double eta, void *p) |
| void | voigtProfile (const vector< double > &x, double sigma, double gamma, vector< double > &y) |
| double | fitVoigtFunc (const gsl_vector *v, void *params) |
| double | fitDoniachSunjic (const gsl_vector *v, void *params) |
| double | fitLikeLogGaussian (const gsl_vector *v, void *params) |
| double | fitExpNorm (const gsl_vector *v, void *params) |
| void | expNormLSQDerivs (const gsl_vector *v, void *params, gsl_vector *df) |
| template<class T > | |
| bool | andersonDarlingStatistic (std::vector< T > vals, float &meanV, float &stdevVal, float &statistic, size_t &undefCount, bool computeMeanAndStdev=true) |
| void | makeHistogram (const vector< float > &data, float start, float end, float step, vector< float > &histVals) |
| void | diff (const vector< float > &in, vector< float > &out) |
| template<class T , class T2 > | |
| std::iterator_traits< T >::value_type | kahansum (T begin, T end, T2 other) |
| double | zechRoot (double sigGuess, void *params) |
| template<class T > | |
| void | cumTrapezoid (const vector< T > &x, const vector< T > &vals, vector< T > &res) |
| double | gauss_ratio_pdf (double x, double muX, double muY, double varX, double varY) |
Variables | |
| LibVersion | libVersion |
| RandNumGen | randGen |
| const char * | RECORDREAD_ERR_STRINGS [] |
| const char * | ATO_ERR_STRINGS [] |
| Human readable error messages for use with ATO reader return values. More... | |
| const char * | OPS_ENUM_ERRSTRINGS [] |
| const unsigned int | NUM_ELEMENTS =119 |
| const size_t | EPOS_RECORD_SIZE = 11*4 |
| const unsigned int | ELEM_SINGLE_NODE_POINT =15 |
| const unsigned int | ELEM_TWO_NODE_LINE =1 |
| const unsigned int | ELEM_THREE_NODE_TRIANGLE =2 |
| const unsigned int | ELEM_FOUR_NODE_TETRAHEDRON =4 |
| const char * | MESH_LOAD_ERRS [] |
| bool | qhullInited =false |
| const unsigned int | HULL_GRAB_SIZE =4096 |
| int | aiCubeEdgeFlags [256] |
| int | a2iTriangleConnectionTable [256][16] |
| const unsigned int | VERTEX_OFFSET [8][3] |
| int | edgeRemap [12] |
| bool | hardAssert =true |
| const size_t | maximumLinearTable [] |
| const char | MULTIRANGE_FORMAT_VERSION [] = "0.0.1" |
| const size_t | MAX_LINE_SIZE = 16536 |
| const size_t | MAX_RANGEFILE_SIZE = 50*1024*1024 |
| const char * | RANGE_EXTS [] |
| const char * | elementList [] |
| const char * | ABUNDANCE_ERROR [] |
| const size_t | PROGRESS_REDUCE =500 |
Typedef Documentation
◆ ATO_ENTRY
| typedef struct AtomProbe::ATO_ENTRY AtomProbe::ATO_ENTRY |
◆ IONHIT
| typedef struct AtomProbe::IONHIT AtomProbe::IONHIT |
Record as stored in a .POS file.
Enumeration Type Documentation
◆ anonymous enum
| anonymous enum |
| Enumerator | |
|---|---|
| HULL_SURFREDUCE_NEGATIVE_SCALE_FACT | |
| HULL_ERR_USER_ABORT | |
| HULL_ERR_NO_MEM | |
Definition at line 26 of file convexHull.h.
◆ anonymous enum
| anonymous enum |
| Enumerator | |
|---|---|
| ATO_OPEN_FAIL | |
| ATO_EMPTY_FAIL | |
| ATO_SIZE_ERR | |
| ATO_VERSIONCHECK_ERR | |
| ATO_MEM_ERR | |
| ATO_BAD_ENDIAN_DETECT | |
| ATO_ENUM_END | |
Definition at line 49 of file dataFiles.h.
◆ anonymous enum
| anonymous enum |
| Enumerator | |
|---|---|
| TEXT_ERR_FILE_OPEN | |
| TEXT_ERR_FILE_NUM_FIELDS | |
| TEXT_ERR_FILE_FORMAT | |
Definition at line 60 of file dataFiles.h.
◆ anonymous enum
| anonymous enum |
Error codes for OPS file loading.
Definition at line 183 of file dataFiles.h.
◆ anonymous enum
| anonymous enum |
| Enumerator | |
|---|---|
| MULTIRANGE_FORMAT_XML | |
Definition at line 57 of file multiRange.h.
◆ anonymous enum
| anonymous enum |
◆ anonymous enum
| anonymous enum |
◆ anonymous enum
| anonymous enum |
composition parsing error messages
| Enumerator | |
|---|---|
| COMPPARSE_BAD_INPUT_FORMAT | |
| COMPPARSE_BAD_COMP_VALUE | |
| COMPPARSE_BAD_BAL_USAGE | |
| COMPPARSE_BAD_SYMBOL | |
| COMPPARSE_DUPLICATE_SYMBOL | |
| COMPPARSE_BAD_TOTAL_COMP | |
Definition at line 26 of file massconvert.h.
◆ anonymous enum
| anonymous enum |
| Enumerator | |
|---|---|
| CRYSTAL_BAD_UNIT_CELL | |
| CRYSTAL_ENUM_END | |
Definition at line 96 of file generate.h.
◆ anonymous enum
| anonymous enum |
| Enumerator | |
|---|---|
| LATTICE_FCC | |
| LATTICE_BCC | |
| LATTICE_HCP | |
| LATTICE_END_OF_ENUM | |
Definition at line 28 of file millerIndex.h.
◆ anonymous enum
| anonymous enum |
◆ anonymous enum
| anonymous enum |
◆ anonymous enum
| anonymous enum |
| Enumerator | |
|---|---|
| EVOLUTION_MODE_SHANKANGLE | |
Definition at line 29 of file reconstruction-simple.h.
◆ anonymous enum
| anonymous enum |
Background fitting modes.
| Enumerator | |
|---|---|
| BACK_FIT_MODE_CONST_TOF | |
| BACK_FIT_MODE_ENUM_END | |
Definition at line 22 of file processing.h.
◆ anonymous enum
| anonymous enum |
| Enumerator | |
|---|---|
| POS_ALLOC_FAIL | |
| POS_OPEN_FAIL | |
| POS_SIZE_MODULUS_ERR | |
| POS_SIZE_EMPTY_ERR | |
| POS_READ_FAIL | |
| POS_NAN_LOAD_ERROR | |
| POS_FILE_ENUM_END | |
Definition at line 53 of file dataFiles.cpp.
◆ anonymous enum
| anonymous enum |
| Enumerator | |
|---|---|
| TAPSIM_FILE_FORMAT_FAIL | |
| TAPSIM_OPEN_FAIL | |
Definition at line 65 of file dataFiles.cpp.
◆ anonymous enum
| anonymous enum |
Definition at line 48 of file multiRange.cpp.
◆ anonymous enum
| anonymous enum |
◆ anonymous enum
| anonymous enum |
◆ anonymous enum
◆ anonymous enum
| anonymous enum |
◆ anonymous enum
| anonymous enum |
◆ anonymous enum
| anonymous enum |
◆ anonymous enum
| anonymous enum |
| Enumerator | |
|---|---|
| PROP_PARSE_ERR | |
| PROP_BAD_ATT | |
Definition at line 36 of file XMLHelper.h.
◆ anonymous enum
| anonymous enum |
| Enumerator | |
|---|---|
| RECORDREAD_ERR_GET_FILESIZE | |
| RECORDREAD_ERR_FILESIZE_MODULO | |
| RECORDREAD_ERR_FILE_OPEN | |
| RECORDREAD_ERR_NOMEM | |
| RECORDREAD_BAD_FILEREAD | |
| RECORDREAD_ERR_CHUNKOFFSET | |
| RECORDREAD_BAD_RECORD | |
Definition at line 34 of file dataFiles.h.
◆ PointDir
| enum AtomProbe::PointDir |
Function Documentation
◆ accumulateCorrelationHistogram()
| bool AtomProbe::accumulateCorrelationHistogram | ( | const std::vector< EPOS_ENTRY > & | eposIons, |
| std::vector< std::vector< unsigned int > > & | histogram, | ||
| float | stepMass, | ||
| float | endMass, | ||
| bool | lowerTriangular = true |
||
| ) |
Increments a correlation histogram, as per correlationHistogram,.
For input parameters, see correlationHistogram(...). This version will have lower peak memory input, as eposIons can be chunked.
- The input histogram must be zero sized, or have the correct size when used. If zero sized, it will be resized and zeroed appropriately to contents
- The default here is that the lower triangular is true, unlike in correlationHistogram(...) so when performing the final accumulation, you may wish to set this to false, to ensure that histogram_ij = histogram_ji.
- Do not change endMass or stepMass between calls on the same histogram
Definition at line 119 of file histogram.cpp.
References incrementCorrelationHist().
Referenced by main().

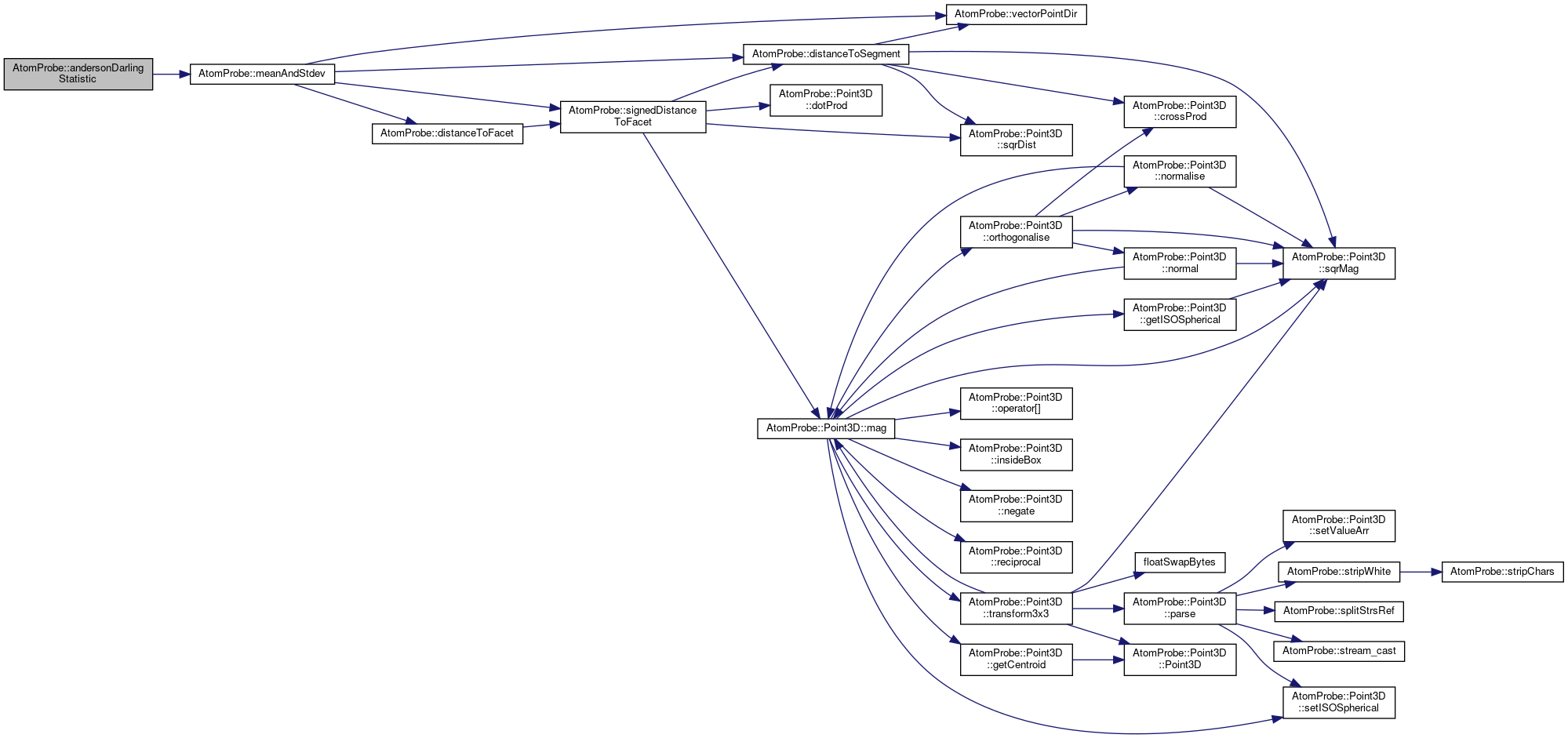
◆ andersonDarlingStatistic()
| bool AtomProbe::andersonDarlingStatistic | ( | std::vector< T > | vals, |
| float & | meanV, | ||
| float & | stdevVal, | ||
| float & | statistic, | ||
| size_t & | undefCount, | ||
| bool | computeMeanAndStdev = true |
||
| ) |
Definition at line 51 of file processing.cpp.
References meanAndStdev().
Referenced by doFitBackground().
◆ antiRotateMatch()
| bool AtomProbe::antiRotateMatch | ( | const unsigned int * | a, |
| const unsigned int * | b, | ||
| size_t | n | ||
| ) |
Definition at line 63 of file mesh.cpp.
Referenced by AtomProbe::TRIANGLE::edgesMismatch().
◆ askAssert()
| void AtomProbe::askAssert | ( | const char * | filename, |
| unsigned int | lineNumber | ||
| ) |
Either abort program, or ask the user what to do for an assertion. depending on hardAssert setting.
Definition at line 35 of file aptAssert.cpp.
◆ buildFrequencyTable()
| void AtomProbe::buildFrequencyTable | ( | const vector< ISOTOPE_ENTRY > & | solutionVec, |
| vector< size_t > & | solutionElements, | ||
| vector< size_t > & | solutionFrequency | ||
| ) |
Definition at line 50 of file filter.cpp.
References ASSERT.
Referenced by filterPeakNeedBiggerObs(), and maxExplainedFraction().
◆ castVoxels()
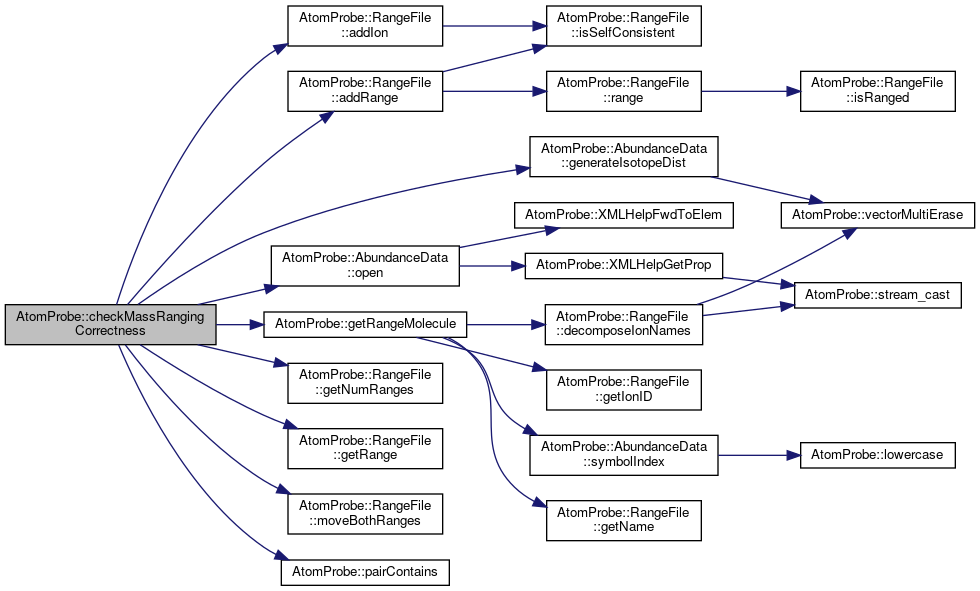
◆ checkMassRangingCorrectness()
| void AtomProbe::checkMassRangingCorrectness | ( | const AtomProbe::RangeFile & | rng, |
| AtomProbe::AbundanceData & | massTable, | ||
| float | massTolerance, | ||
| unsigned int | maxChargeState, | ||
| unsigned int | maxComponents, | ||
| std::vector< bool > & | badRanges | ||
| ) |
Ensure that each mass given spans a peak that should exist.
Each entry in badRanges is true if an inconsistent range is detected. There will be one entry per range in the parent rangefile Some ranges may be skipped if they cannot be understood maxComponents will skip checks for ions with more than this many atoms in the ionic molecule. Eg H2 would not be checked if maxComponents=1
Definition at line 101 of file rangeCheck.cpp.
References AtomProbe::RangeFile::addIon(), AtomProbe::RangeFile::addRange(), AtomProbe::RGBf::blue, AtomProbe::RANGE_MOLECULE::components, AtomProbe::AbundanceData::generateIsotopeDist(), AtomProbe::RangeFile::getNumRanges(), AtomProbe::RangeFile::getRange(), getRangeMolecule(), AtomProbe::RGBf::green, AtomProbe::RANGE_MOLECULE::isOK, AtomProbe::RangeFile::moveBothRanges(), AtomProbe::AbundanceData::open(), pairContains(), AtomProbe::RGBf::red, and TEST.
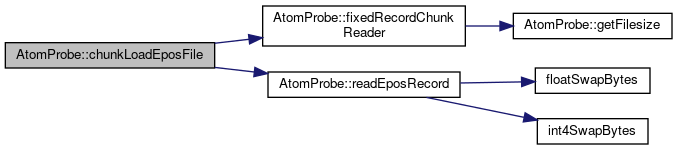
◆ chunkLoadEposFile()
| size_t AtomProbe::chunkLoadEposFile | ( | std::vector< EPOS_ENTRY > & | outData, |
| const char * | filename, | ||
| unsigned int | chunkSize, | ||
| unsigned int | chunkOffset, | ||
| unsigned int & | nEntriesLeft | ||
| ) |
Load an "EPOS" file, with a maximum chunk size.
This is useful to avoid out-of-memory situations when working with large datasets If the file has fewer EPOS entries left than the chunk size, only the remaining entries will be read. Increment chunkOffset to seek to the next part of the file The file will be re-opened each time, so larger chunk sizes will increase speed at the cost of memory.
outData : the output to write the epos data to filename : the name of the EPOS formatted file to open chunkSize : the maximum number of entries to read in one go chunkOffset : the position (in chunks) of the file to start reading from, the caller must increment this to prevent the same section being re-read nEntriesLeft : returns the number of entries left in the file, The return value is nonzero on error, and is part of the RECORDREAD_ERR enum
Definition at line 765 of file dataFiles.cpp.
References EPOS_RECORD_SIZE, fixedRecordChunkReader(), and readEposRecord().
Referenced by getAtoErrString(), and main().
◆ computeComposition()
| void AtomProbe::computeComposition | ( | const std::vector< unsigned int > & | countData, |
| std::vector< float > & | compositionData | ||
| ) |
Definition at line 29 of file composition.cpp.
Referenced by main().
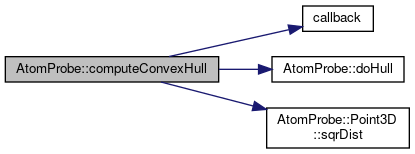
◆ computeConvexHull()
| unsigned int AtomProbe::computeConvexHull | ( | const std::vector< IonHit > & | data, |
| unsigned int * | progress, | ||
| bool(*)(bool) | callback, | ||
| std::vector< Point3D > & | curHull, | ||
| bool | freeHull | ||
| ) |
Obtain the convex hull of a set of ions.
An existing set of points on the convex hull, "curHull", can be given as a starting set, optionally. For the last call to this routine , set "freeHull" to true. Returns 0 on success, nonzero on error, with error codes in above enum
Definition at line 149 of file convexHull.cpp.
References callback(), doHull(), HULL_ERR_NO_MEM, HULL_ERR_USER_ABORT, PROGRESS_REDUCE, and AtomProbe::Point3D::sqrDist().
Referenced by GetReducedHullPts().
◆ computeIonDistAdjacency()
| void AtomProbe::computeIonDistAdjacency | ( | const MultiRange & | mrf, |
| const AbundanceData & | abundance, | ||
| const OVERLAP_PROBLEM_SETTINGS & | settings, | ||
| gsl_matrix *& | m, | ||
| gsl_vector *& | vOwnership, | ||
| gsl_vector *& | vMass | ||
| ) |
Calculate the adjacency matrix for the ions in a multirange.
Uses tolerance and other data in settings. Range data is NOT used. Vectors and matrices should not be allocated, but must be freed using gsl_matrix_free and gsl_vector_free after use. Return values are a 0/1 adjacency matrix, and two vectors, one describing the owner index of each ion and the other the mass. return matrix will be square, and vectors will have same size as matrix
Definition at line 40 of file componentAnalysis.cpp.
References ASSERT, AtomProbe::AbundanceData::generateIsotopeDist(), AtomProbe::MultiRange::getMolecule(), AtomProbe::MultiRange::getNumIons(), AtomProbe::OVERLAP_PROBLEM_SETTINGS::intensityTolerance, AtomProbe::OVERLAP_PROBLEM_SETTINGS::massTolerance, AtomProbe::OVERLAP_PROBLEM_SETTINGS::maxDefaultCharge, AtomProbe::AbundanceData::symbolIdxFromAtomicNumber(), and vectorMultiErase().
Referenced by computeRangeAdjacency().
◆ computeRangeAdjacency()
| void AtomProbe::computeRangeAdjacency | ( | const MultiRange & | mrf, |
| const OVERLAP_PROBLEM_SETTINGS & | settings, | ||
| gsl_matrix *& | m | ||
| ) |
Definition at line 172 of file componentAnalysis.cpp.
References AtomProbe::MultiRange::addIon(), AtomProbe::MultiRange::addRange(), ASSERT, AtomProbe::SIMPLE_SPECIES::atomicNumber, AtomProbe::RGBf::blue, computeIonDistAdjacency(), AtomProbe::SIMPLE_SPECIES::count, AtomProbe::AbundanceData::getAtomicNumber(), AtomProbe::MultiRange::getNumRanges(), AtomProbe::MultiRange::getRange(), AtomProbe::RGBf::green, gsl_print_matrix(), AtomProbe::OVERLAP_PROBLEM_SETTINGS::intensityTolerance, AtomProbe::OVERLAP_PROBLEM_SETTINGS::massTolerance, AtomProbe::OVERLAP_PROBLEM_SETTINGS::maxDefaultCharge, AtomProbe::AbundanceData::open(), rangeOverlaps(), AtomProbe::RGBf::red, AtomProbe::AbundanceData::symbolIndex(), TEST, and WARN.
◆ computeRotationMatrix() [1/2]
| void AtomProbe::computeRotationMatrix | ( | const Point3D & | ur1, |
| const Point3D & | ur2, | ||
| const Point3D & | r1, | ||
| const Point3D & | r2, | ||
| gsl_matrix * | m | ||
| ) |
Given two orthogonal and normal basis vectors, r1 and r2, and two matching rotated vectors ur1 and ur2, compute the rotation matrix that will transform between them.
i.e. r = M*ur, vector pairs (ur1,ur2) and (r1,r2) should be orthogonal and normalised matrix m must be pre-allocated as a 3x3 matrix, using e.g gsl_matrix_alloc(3,3)
Definition at line 83 of file rotations.cpp.
References ASSERT, AtomProbe::Point3D::crossProd(), gsl_determinant(), and AtomProbe::Point3D::sqrMag().
Referenced by computeRotationMatrix(), getRotationMatrix(), and main().
◆ computeRotationMatrix() [2/2]
| void AtomProbe::computeRotationMatrix | ( | const Point3D & | ur1, |
| const Point3D & | ur2, | ||
| const Point3D & | r1, | ||
| const Point3D & | r2, | ||
| std::vector< std::vector< float > > & | m | ||
| ) |
Convenience wrapper for conputeRotationMatrix, which uses vector containers.
output vectors will be 3x3, indexed vector[row][column]. Presizing is not required
Definition at line 67 of file rotations.cpp.
References computeRotationMatrix().
◆ computeRotationMatrixWahba()
| unsigned int AtomProbe::computeRotationMatrixWahba | ( | const std::vector< Point3D > & | unrotated, |
| const std::vector< Point3D > & | rotated, | ||
| const std::vector< float > & | weights, | ||
| gsl_matrix *& | R | ||
| ) |
Given a series of paired observations of directions, compute the least squares rotation matrix between them.
This computes the least-squares rotation matrix that transforms the unrotated points to the rotated ones. rotated points and unrotated points must be the same size, must have >=2 entries, and must not be degenerate. Each point must be normalised. Weights is a series of weighting factor (e.g. inverse error) for each point. If R is nullptr on input, then it will be replaced with a matrix allocated by gsl_matrix_alloc(). The caller should deallocate this with gsl_matrix_free() after use.
Definition at line 147 of file rotations.cpp.
References ASSERT, gsl_determinant(), and gsl_matrix_mult().
Referenced by getRotationMatrix().
◆ convertEPOStoPos()
| void AtomProbe::convertEPOStoPos | ( | const std::vector< EPOS_ENTRY > & | eposEntry, |
| std::vector< IonHit > & | posFile | ||
| ) |
Convert an incoming entry of EPOS files to pos, and the append this to the pos vector given.
Definition at line 29 of file convert.cpp.
◆ convertFileStringToCanonical()
| std::string AtomProbe::convertFileStringToCanonical | ( | const std::string & | s | ) |
Convert a path format into a unix path from native format.
Referenced by strAppend().
◆ convertFileStringToNative()
| std::string AtomProbe::convertFileStringToNative | ( | const std::string & | s | ) |
Convert a path format into a native path from unix format.
Referenced by strAppend().
◆ convertMassToMol()
| void AtomProbe::convertMassToMol | ( | const AbundanceData & | atomTable, |
| const std::map< unsigned int, double > & | compositionMass, | ||
| std::map< unsigned int, double > & | compositionMols | ||
| ) |
Convert the given composition from mass fraction data to molar fraction.
inputs are: compositionMass - relative mass fractions (this will be normalised automatically) massValues - weights of the masses input compositionMols - output results
Definition at line 74 of file massconvert.cpp.
References AtomProbe::AbundanceData::getNominalMass().
Referenced by parseCompositionData().
◆ convertMolToMass()
| void AtomProbe::convertMolToMass | ( | const AbundanceData & | atomTable, |
| const std::map< unsigned int, double > & | compositionMols, | ||
| std::map< unsigned int, double > & | compositionMass | ||
| ) |
Convert the given composition from molar fraction data to mass fraction.
Inputs are:
- atomTable - the natural abundance table, to provide weights
- compositionMols : relative molar fractions (this will be normalised automatically). First entry in map is ion index in abundance table. Second is relative fraction.
Outputs are:
- compositionMass : output results, as mass fractions First entry is the position in the abundance table, second is relative fraction
Definition at line 41 of file massconvert.cpp.
References ASSERT, and AtomProbe::AbundanceData::getNominalMass().
Referenced by parseCompositionData().
◆ correlationHistogram()
| bool AtomProbe::correlationHistogram | ( | const std::vector< EPOS_ENTRY > & | eposIons, |
| std::vector< std::vector< unsigned int > > & | histogram, | ||
| float | stepMass, | ||
| float | endMass, | ||
| bool | lowerTriangular = false |
||
| ) |
Generates an ion "correlation histogram" from a vector of EPOS ions. The input vector MUST
The nature of the plot is described in the following paper (Saxey et al, 2011): DOI : 10.1016/j.ultramic.2010.11.021, see e.g, Figure 3a.
The histogram will be allocated for you - do not pre-allocate
As the plot is symmetric around the m1=m2 line, the lowerTriangular setting controls if mirroring of the calculation is done. Setting this to true will ensure that only the lower half of the data field is calculated (faster).
StepMass > 0 EndMass > 0
in both cases, the histogram will use the full rectangular memory space (arrays will not be ragged)
Definition at line 85 of file histogram.cpp.
References incrementCorrelationHist().

◆ countIntensityEvents()
| unsigned int AtomProbe::countIntensityEvents | ( | const vector< pair< float, float > > & | data, |
| float | minV, | ||
| float | maxV | ||
| ) |
Definition at line 36 of file filter.cpp.
Referenced by maxExplainedFraction().
◆ createMassBackground()
| void AtomProbe::createMassBackground | ( | float | massStart, |
| float | massEnd, | ||
| unsigned int | nBinsMass, | ||
| float | tofBackIntensity, | ||
| std::vector< float > & | histogram | ||
| ) |
Build a histogram of the background counts.
- Inputs : Start and end mass for background window, and bin count
- tofBackIntensity is the intensity level per unit time (proportional to sqrt mass) in the background, as obtained by doFitBackground.
Note : You may need to divide by bin width, depending on your application
Definition at line 224 of file processing.cpp.
Referenced by findPeaks().
◆ cumTrapezoid()
| void AtomProbe::cumTrapezoid | ( | const vector< T > & | x, |
| const vector< T > & | vals, | ||
| vector< T > & | res | ||
| ) |
Definition at line 461 of file confidence.cpp.
References ASSERT, and kahansum().

◆ det3by3()
| double AtomProbe::det3by3 | ( | const double * | ptArray | ) |
Definition at line 256 of file misc.cpp.
Referenced by dotProduct(), and pyramidVol().
◆ Determinant()
| float AtomProbe::Determinant | ( | float ** | a, |
| int | n | ||
| ) |
Definition at line 172 of file mesh.cpp.
References ASSERT, and signVal().
Referenced by fourDeterminant().
◆ diff()
| void AtomProbe::diff | ( | const vector< float > & | in, |
| vector< float > & | out | ||
| ) |
Definition at line 249 of file processing.cpp.
Referenced by findPeaks().
◆ digitString()
| std::string AtomProbe::digitString | ( | unsigned int | thisDigit, |
| unsigned int | maxDigit | ||
| ) |
Generate a string with leading digits up to maxDigit (eg, if maxDigit is 424, and thisDigit is 1.
Definition at line 115 of file stringFuncs.cpp.
References stream_cast().
Referenced by strAppend().

◆ distanceToFacet()
| float AtomProbe::distanceToFacet | ( | const Point3D & | fA, |
| const Point3D & | fB, | ||
| const Point3D & | fC, | ||
| const Point3D & | normal, | ||
| const Point3D & | p | ||
| ) |
Definition at line 250 of file misc.cpp.
References signedDistanceToFacet().
Referenced by GetReducedHullPts(), and meanAndStdev().
◆ distanceToSegment()
Definition at line 156 of file misc.cpp.
References AtomProbe::Point3D::crossProd(), POINTDIR_TOGETHER, AtomProbe::Point3D::sqrDist(), AtomProbe::Point3D::sqrMag(), and vectorPointDir().
Referenced by meanAndStdev(), and signedDistanceToFacet().
◆ doFitBackground()
| unsigned int AtomProbe::doFitBackground | ( | const std::vector< float > & | massData, |
| BACKGROUND_PARAMS & | params | ||
| ) |
Perform a background fit, assuming constant TOF noise, from 0->inf time.
- Background params has the input and output data for the fit.
dataIn requires raw mass data, not histogram counts.
- returns zero on success, nonzero on error Return value is stored in params.intensity, as counts/bin. Use createMassBackground to re-generate the histogram
Definition at line 160 of file processing.cpp.
References andersonDarlingStatistic(), ASSERT, AtomProbe::BACKGROUND_PARAMS::binWidth, AtomProbe::BACKGROUND_PARAMS::FIT_FAIL_AVG_COUNTS, AtomProbe::BACKGROUND_PARAMS::FIT_FAIL_DATA_NON_GAUSSIAN, AtomProbe::BACKGROUND_PARAMS::FIT_FAIL_INSUFF_DATA, AtomProbe::BACKGROUND_PARAMS::FIT_FAIL_MIN_REQ_BINS, AtomProbe::BACKGROUND_PARAMS::intensity, makeHistogram(), AtomProbe::BACKGROUND_PARAMS::massEnd, AtomProbe::BACKGROUND_PARAMS::massStart, and AtomProbe::BACKGROUND_PARAMS::stdev.
◆ doHull()
| unsigned int AtomProbe::doHull | ( | unsigned int | bufferSize, |
| double * | buffer, | ||
| vector< Point3D > & | resHull, | ||
| Point3D & | midPoint, | ||
| bool | freeHullOnExit | ||
| ) |
Definition at line 53 of file convexHull.cpp.
References HULL_ERR_NO_MEM.
Referenced by computeConvexHull().
◆ doniachSunjic()
| void AtomProbe::doniachSunjic | ( | const std::vector< double > & | x, |
| double & | a, | ||
| double & | F, | ||
| double & | mu, | ||
| double & | amp, | ||
| std::vector< double > & | y | ||
| ) |
generate a Doniach-Sunjic profile
Definition at line 502 of file fitting.cpp.
References M_PI.
Referenced by fitDoniachSunjic().
◆ dotProduct()
|
inline |
Inline func for calculating a(dot)b.
Definition at line 128 of file misc.h.
References det3by3(), pyramidVol(), and triIsDegenerate().
Referenced by GetReducedHullPts().
◆ edgeIdx()
| unsigned int AtomProbe::edgeIdx | ( | unsigned int | i, |
| unsigned int | j | ||
| ) |
◆ estimateRank()
| unsigned int AtomProbe::estimateRank | ( | const gsl_matrix * | m, |
| float | tolerance = sqrt(std::numeric_limits<float>::epsilon()) |
||
| ) |
Estimate the rank of the given matrix.
Definition at line 58 of file misc.cpp.
Referenced by leastSquaresOverlapSolve(), and pyramidVol().
◆ expNorm()
| void AtomProbe::expNorm | ( | const std::vector< double > & | x, |
| double & | K, | ||
| double & | mu, | ||
| double & | sigma, | ||
| double & | amp, | ||
| std::vector< double > & | y | ||
| ) |
Exponentially decaying normal distribution.
Definition at line 532 of file fitting.cpp.
Referenced by fitExpNorm(), and main().
◆ expNormLSQDerivs()
| void AtomProbe::expNormLSQDerivs | ( | const gsl_vector * | v, |
| void * | params, | ||
| gsl_vector * | df | ||
| ) |
Definition at line 551 of file fitting.cpp.
References M_PI, AtomProbe::DATA_FIT::x, and AtomProbe::DATA_FIT::y.
◆ filterBySolutionPPM()
| void AtomProbe::filterBySolutionPPM | ( | const AtomProbe::AbundanceData & | massTable, |
| float | minPpm, | ||
| std::vector< std::vector< AtomProbe::ISOTOPE_ENTRY > > & | solutions | ||
| ) |
Use the maximum possible PPM for each isotopic combination to filter possible solutions.
For example, 17^O:17^O:17^O in a naturally abundant mix has a maximum possible concentration of 5e-11 at. fraction. Isotope group tolerance is used to group the abundance distribution of nearby isotopes
massTable - a table of natural abundances minPpm - Minimum PPM below which to discard solutions solutions - list of solutions, which will be culled.
Definition at line 83 of file filter.cpp.
References AtomProbe::ISOTOPE_ENTRY::abundance, AtomProbe::AbundanceData::isotope(), and vectorMultiErase().
◆ filterPeakNeedBiggerObs()
| void AtomProbe::filterPeakNeedBiggerObs | ( | const AtomProbe::AbundanceData & | massTable, |
| const std::vector< float > & | peakData, | ||
| float | tolerance, | ||
| size_t | solutionCharge, | ||
| std::vector< std::vector< AtomProbe::ISOTOPE_ENTRY > > & | solutions | ||
| ) |
Remove peaks for which an experimental set of observed peaks do not show a peak at a position which is expected from the peak distribution for this element. Only works with single-atom peaks
- Solutions are the input solutions,
- peakData is a list of peak mass positions
- tolerance is the error allowed to fit a peak (+/-)
- solutionCharge - charge state for the input isotope entries
Definition at line 119 of file filter.cpp.
References ASSERT, buildFrequencyTable(), AtomProbe::AbundanceData::generateIsotopeDist(), and vectorMultiErase().
◆ findMaxLessThanOrEq()
| size_t AtomProbe::findMaxLessThanOrEq | ( | const vector< std::pair< size_t, size_t > > & | v, |
| size_t | value | ||
| ) |
◆ findNearVerticies() [1/2]
| void AtomProbe::findNearVerticies | ( | float | tolerance, |
| const vector< Point3D > & | ptVec, | ||
| vector< std::pair< size_t, vector< size_t > > > & | clusterList | ||
| ) |
Definition at line 365 of file mesh.cpp.
References ASSERT.
Referenced by AtomProbe::Mesh::mergeDuplicateVertices(), and AtomProbe::Mesh::setTriangleMesh().
◆ findNearVerticies() [2/2]
| void AtomProbe::findNearVerticies | ( | float | tolerance, |
| const vector< Point3D > & | ptVec, | ||
| std::list< std::pair< size_t, vector< size_t > > > & | clusterList | ||
| ) |
◆ findOverlaps() [1/4]
| void AtomProbe::findOverlaps | ( | const AbundanceData & | natData, |
| const RangeFile & | rng, | ||
| float | massDelta, | ||
| unsigned int | maxCharge, | ||
| std::vector< std::pair< size_t, size_t > > & | overlapIdx, | ||
| std::vector< std::pair< float, float > > & | overlapMasses | ||
| ) |
Find the overlaps stemming from a given rangefile.
An overlap is defined as "natural abundances within +-massDelta at same mass to charge"
- Species not in the rangefile will not be marked as overlapping
- FIXME: The output index is abused at this time, the range's index is actually given by dividing the output by the mass to charge. The charge state is given by modulo (massToCharge) +1
- The range mass information in the rangefile is not used - it is abused as an ion list
Inputs are:
- rng: The rangefile to use. The species in the rangefile are used to compute the overlaps
- massDelta: tolerance in amu,
- maxCharge: the maximum charge that species can be estimated to have
Outputs are:
- overlapIdx: the range indices of the overlapping species (ion index in rangefile)
- overlapMasses: the masses of the overlapping species, which should be within +-massDelta of one another
Note that ion's names will be decomposed, so Fe2H will be considered an Fe-Fe-H molecule
Referenced by findOverlaps(), and main().
◆ findOverlaps() [2/4]
| void AtomProbe::findOverlaps | ( | const AbundanceData & | natData, |
| const std::vector< std::string > & | ions, | ||
| float | massDelta, | ||
| unsigned int | maxCharge, | ||
| std::vector< std::pair< size_t, size_t > > & | overlapIdx, | ||
| std::vector< std::pair< float, float > > & | overlapMasses | ||
| ) |
As per findOverlaps(...Rangefile ...) , but uses a vector of ions instead.
◆ findOverlaps() [3/4]
| void AtomProbe::findOverlaps | ( | const AbundanceData & | natData, |
| const RangeFile & | rng, | ||
| float | massDelta, | ||
| unsigned int | maxCharge, | ||
| vector< pair< size_t, size_t > > & | overlapIdx, | ||
| vector< pair< float, float > > & | overlapMasses | ||
| ) |
Definition at line 94 of file overlaps.cpp.
References ASSERT, AtomProbe::RangeFile::decomposeIonById(), findOverlapsFromSpectra(), AtomProbe::AbundanceData::generateGroupedIsotopeDist(), AtomProbe::AbundanceData::getAtomicNumber(), AtomProbe::RangeFile::getNumIons(), AtomProbe::AbundanceData::getSymbolIndices(), and vectorMultiErase().
◆ findOverlaps() [4/4]
| void AtomProbe::findOverlaps | ( | const AbundanceData & | natData, |
| const vector< string > & | ionNames, | ||
| float | massDelta, | ||
| unsigned int | maxCharge, | ||
| vector< pair< size_t, size_t > > & | overlapIdx, | ||
| vector< pair< float, float > > & | overlapMasses | ||
| ) |
Definition at line 218 of file overlaps.cpp.
References AtomProbe::RangeFile::addIon(), AtomProbe::RangeFile::addRange(), AtomProbe::RGBf::blue, findOverlaps(), AtomProbe::RGBf::green, AtomProbe::AbundanceData::open(), AtomProbe::RGBf::red, and TEST.
◆ findOverlapsFromSpectra() [1/2]
| void AtomProbe::findOverlapsFromSpectra | ( | const vector< vector< pair< float, float > > > & | massDistributions, |
| float | massDelta, | ||
| const set< unsigned int > & | skipIDs, | ||
| vector< pair< size_t, size_t > > & | overlapIdx, | ||
| vector< pair< float, float > > & | overlapMasses | ||
| ) |
Definition at line 46 of file overlaps.cpp.
References EQ_TOLV.
◆ findOverlapsFromSpectra() [2/2]
| void AtomProbe::findOverlapsFromSpectra | ( | const std::vector< std::vector< std::pair< float, float > > > & | massDistributions, |
| float | massDelta, | ||
| const std::set< unsigned int > & | skipIDs, | ||
| std::vector< std::pair< size_t, size_t > > & | overlapIdx, | ||
| std::vector< std::pair< float, float > > & | overlapMasses | ||
| ) |
Find overlaps in the given mass distributions, as per findOverlaps(...Rangefile...)
Referenced by findOverlaps().
◆ findPeaks()
| void AtomProbe::findPeaks | ( | const std::vector< float > & | x0, |
| std::vector< unsigned int > & | peakInds, | ||
| float | sel, | ||
| bool | autoSel = true, |
||
| bool | includeEndpoints = true |
||
| ) |
Simple peak-finding algorithm.
Adapted from Nathaniel Yoder's: https://uk.mathworks.com/matlabcentral/fileexchange/25500-peakfinder-x0-sel-thresh-extrema-includeendpoints-interpolate which is BSD licenced.
The performance of this function may or may not be suitable for your application, and your histogram may require pre-processing to optimise false postive/false-negative performance
- x0 : input mass histogram, must be > 2 elements
- sel : Selectivity for peaks - larger is more selective
- autosel : whether to determine sel automatically, or not
- includeEndpoints : allow edge values to be peaks
Definition at line 268 of file processing.cpp.
References ASSERT, createMassBackground(), diff(), makeHistogram(), NUM_IONS, and selectElements().
◆ fitDoniachSunjic() [1/2]
| bool AtomProbe::fitDoniachSunjic | ( | const std::vector< double > & | x, |
| const std::vector< double > & | y, | ||
| double & | a, | ||
| double & | f, | ||
| double & | mu, | ||
| double & | amp, | ||
| bool | autoInit = true |
||
| ) |
Fit a Doniach-Sunjic curve.
Definition at line 204 of file fitting.cpp.
References weightedMean(), AtomProbe::DATA_FIT::x, and AtomProbe::DATA_FIT::y.

◆ fitDoniachSunjic() [2/2]
| double AtomProbe::fitDoniachSunjic | ( | const gsl_vector * | v, |
| void * | params | ||
| ) |
Definition at line 60 of file fitting.cpp.
References doniachSunjic(), lsq(), AtomProbe::DATA_FIT::x, and AtomProbe::DATA_FIT::y.
◆ fitExpNorm() [1/2]
| bool AtomProbe::fitExpNorm | ( | const std::vector< double > & | x, |
| const std::vector< double > & | y, | ||
| double & | K, | ||
| double & | mu, | ||
| double & | sigma, | ||
| double & | amp, | ||
| bool | autoInit = true |
||
| ) |
Fit a smoothed log-gaussian curve (arxiv:0711.4449)
Definition at line 280 of file fitting.cpp.
References weightedMean(), AtomProbe::DATA_FIT::x, and AtomProbe::DATA_FIT::y.
Referenced by main().

◆ fitExpNorm() [2/2]
| double AtomProbe::fitExpNorm | ( | const gsl_vector * | v, |
| void * | params | ||
| ) |
Definition at line 103 of file fitting.cpp.
References expNorm(), lsq(), AtomProbe::DATA_FIT::x, and AtomProbe::DATA_FIT::y.

◆ fitLikeLogGaussian() [1/2]
| bool AtomProbe::fitLikeLogGaussian | ( | const std::vector< double > & | x, |
| const std::vector< double > & | y, | ||
| double & | lambda, | ||
| double & | sigma, | ||
| double & | xp, | ||
| double & | amp, | ||
| double & | h, | ||
| bool | autoInit = true |
||
| ) |
Fit a smoothed log-gaussian curve (arxiv:0711.4449)
Definition at line 360 of file fitting.cpp.
References weightedMean(), AtomProbe::DATA_FIT::x, and AtomProbe::DATA_FIT::y.

◆ fitLikeLogGaussian() [2/2]
| double AtomProbe::fitLikeLogGaussian | ( | const gsl_vector * | v, |
| void * | params | ||
| ) |
Definition at line 80 of file fitting.cpp.
References likeLogGaussian(), lsq(), AtomProbe::DATA_FIT::x, and AtomProbe::DATA_FIT::y.
◆ fitVoigt()
| bool AtomProbe::fitVoigt | ( | const std::vector< double > & | x, |
| const std::vector< double > & | y, | ||
| double & | sigma, | ||
| double & | gamma, | ||
| double & | mu, | ||
| double & | amp, | ||
| bool | autoInit = true |
||
| ) |
Fit a Voigt function to the given X/Y values. Internally, a function minimiser is used.
Voigt functions are used in spectroscopy as a general curve for fitting spectra. they are symmetric, and won't fit "tailed" curves well
autoinit : If true, then initial values are chosen automatically and heuristically, if false, input parameters must be given, and are used to seed the minimiser outputs are sigma/gamma (voigt parameters), mu (x shift) and amp (y-scale)
Definition at line 123 of file fitting.cpp.
References fitVoigtFunc(), weightedMean(), AtomProbe::DATA_FIT::x, and AtomProbe::DATA_FIT::y.
Referenced by main().
◆ fitVoigtFunc()
| double AtomProbe::fitVoigtFunc | ( | const gsl_vector * | v, |
| void * | params | ||
| ) |
Definition at line 44 of file fitting.cpp.
References lsq(), voigtProfile(), AtomProbe::DATA_FIT::x, and AtomProbe::DATA_FIT::y.
Referenced by fitVoigt().
◆ fixedRecordChunkReader()
| unsigned int AtomProbe::fixedRecordChunkReader | ( | const char * | filename, |
| bool(*)(const char *bufRead, const char *destBuf) | recordReader, | ||
| size_t | recordSize, | ||
| std::vector< T > & | outputData, | ||
| unsigned int | chunkSize, | ||
| unsigned int | chunkOffset, | ||
| unsigned int & | nEntriesLeft | ||
| ) |
Definition at line 144 of file dataFiles.cpp.
References getFilesize(), RECORDREAD_BAD_FILEREAD, RECORDREAD_BAD_RECORD, RECORDREAD_ERR_CHUNKOFFSET, RECORDREAD_ERR_FILE_OPEN, RECORDREAD_ERR_FILESIZE_MODULO, RECORDREAD_ERR_GET_FILESIZE, and RECORDREAD_ERR_NOMEM.
Referenced by chunkLoadEposFile().
◆ fixedRecordReader()
| unsigned int AtomProbe::fixedRecordReader | ( | const char * | filename, |
| bool(*)(const char *bufRead, const char *destBuf) | recordReader, | ||
| size_t | recordSize, | ||
| std::vector< T > & | outputData | ||
| ) |
Definition at line 93 of file dataFiles.cpp.
References getFilesize(), RECORDREAD_BAD_FILEREAD, RECORDREAD_BAD_RECORD, RECORDREAD_ERR_FILE_OPEN, RECORDREAD_ERR_FILESIZE_MODULO, RECORDREAD_ERR_GET_FILESIZE, and RECORDREAD_ERR_NOMEM.
Referenced by loadEposFile().

◆ fourDeterminant()
| float AtomProbe::fourDeterminant | ( | const Point3D & | a, |
| const Point3D & | b, | ||
| const Point3D & | c, | ||
| const Point3D & | d | ||
| ) |
Definition at line 212 of file mesh.cpp.
References Determinant().

◆ fpeek()
|
inline |
Definition at line 69 of file helpFuncs.h.
Referenced by AtomProbe::RangeFile::rangeTypeString().
◆ freeConvexHull()
| void AtomProbe::freeConvexHull | ( | ) |
Definition at line 47 of file convexHull.cpp.
Referenced by GetReducedHullPts().
◆ gauss_ratio_pdf()
| double AtomProbe::gauss_ratio_pdf | ( | double | x, |
| double | muX, | ||
| double | muY, | ||
| double | varX, | ||
| double | varY | ||
| ) |
Definition at line 483 of file confidence.cpp.
References M_PI.
◆ gcd()
| int AtomProbe::gcd | ( | int | a, |
| int | b | ||
| ) |
Definition at line 28 of file millerIndex.cpp.
Referenced by AtomProbe::MILLER_TRIPLET::simplify().
◆ genColString() [1/2]
| void AtomProbe::genColString | ( | unsigned char | r, |
| unsigned char | g, | ||
| unsigned char | b, | ||
| unsigned char | a, | ||
| std::string & | s | ||
| ) |
Referenced by strAppend(), and AtomProbe::RGBf::toHex().
◆ genColString() [2/2]
| void AtomProbe::genColString | ( | unsigned char | r, |
| unsigned char | g, | ||
| unsigned char | b, | ||
| std::string & | s | ||
| ) |
Definition at line 308 of file stringFuncs.cpp.
References ucharToHexStr().

◆ generate1DAxialDistHist()
| unsigned int AtomProbe::generate1DAxialDistHist | ( | const std::vector< Point3D > & | pointList, |
| K3DTreeExact & | tree, | ||
| const Point3D & | axisDir, | ||
| float | distMax, | ||
| std::vector< unsigned int > & | histogram | ||
| ) |
Generate a 1D axial distribution function,.
This generates a histogram of point-point distances, from the points in the point list to the points in the tree, within a specified radius of the source point.
This is variously called a 1D "Spatial Distribution Map (SDM)" "Atom Vicinity" or similar
- pointList : the source points for the distribution function
- tree : a pre-generated KD tree with the target points
- distMax : radius of the points around which to search
- histogram : a vector with the number of bins that are required for output. Does not need to be zero filled
Definition at line 37 of file axialdf.cpp.
References ASSERT, AtomProbe::K3DTreeExact::getPtRef(), AtomProbe::K3DTreeExact::ptsInSphere(), AtomProbe::BoundCube::setBounds(), AtomProbe::Point3D::sqrDist(), and AtomProbe::Point3D::sqrMag().
◆ generate1DAxialDistHistSweep()
| unsigned int AtomProbe::generate1DAxialDistHistSweep | ( | const std::vector< Point3D > & | pointList, |
| K3DTreeExact & | tree, | ||
| float | distMax, | ||
| float | dTheta, | ||
| float | dPhi, | ||
| AtomProbe::ProgressBar & | prog, | ||
| std::vector< std::vector< std::vector< unsigned int > > > & | histogram | ||
| ) |
Generate a series of 1D distribution functions, one per pixel in a 2D grid of spherical coordinate directions.
Using a stepped spherical coordinates in equal delta-Theta, delta-Phi increments pointList : the source points from which to search tree : K3D Tree of target points distMax : maximum distance around which to search dTheta : step size in theta (radiians). dPhi : step size in phi (radiians) prog : Progress bar histogram : resultant data as a 2D vector, one bin per angular entry. Each vector will have the same size. The inner most vector contains the analysis data, over +-distMax
Definition at line 131 of file axialdf.cpp.
References ASSERT, AtomProbe::Point3D::dotProd(), AtomProbe::K3DTreeExact::getPtRef(), M_PI, AtomProbe::K3DTreeExact::ptsInSphere(), AtomProbe::BoundCube::setBounds(), AtomProbe::Point3D::setISOSpherical(), AtomProbe::K3DTreeExact::size(), AtomProbe::Point3D::sqrDist(), and AtomProbe::ProgressBar::update().
◆ getAtoErrString()
|
inline |
Definition at line 82 of file dataFiles.h.
References chunkLoadEposFile(), getPosFileErrString(), loadEposFile(), loadPosFile(), loadTextData(), AtomProbe::ATO_ENTRY::mass, savePosFile(), and saveTapsimBin().
◆ getFilesize()
| bool AtomProbe::getFilesize | ( | const char * | fname, |
| size_t & | size | ||
| ) |
Definition at line 107 of file helpFuncs.cpp.
Referenced by fixedRecordChunkReader(), fixedRecordReader(), AtomProbe::RangeFile::openFormat(), AtomProbe::ComparePairFirstReverse::operator()(), and readPosapOps().
◆ getFitErrorMsg()
| string AtomProbe::getFitErrorMsg | ( | unsigned int | errCode | ) |
Obtain a human readable error, which has arison when using doFitBackground Input is the error code from doFitBackground
Definition at line 146 of file processing.cpp.
References ASSERT, and AtomProbe::BACKGROUND_PARAMS::FIT_FAIL_END.
◆ getHardAssert()
| bool AtomProbe::getHardAssert | ( | ) |
Definition at line 25 of file aptAssert.cpp.
References hardAssert.
◆ getPosFileErrString()
| const char * AtomProbe::getPosFileErrString | ( | unsigned int | errMesg | ) |
Definition at line 238 of file dataFiles.cpp.
References ARRAYSIZE, and POS_FILE_ENUM_END.
Referenced by getAtoErrString(), and main().
◆ getRangeMolecule()
| RANGE_MOLECULE AtomProbe::getRangeMolecule | ( | const AbundanceData & | massTable, |
| const RangeFile & | rng, | ||
| unsigned int | rangeId | ||
| ) |
Definition at line 55 of file rangeCheck.cpp.
References AtomProbe::RANGE_MOLECULE::components, AtomProbe::RangeFile::decomposeIonNames(), AtomProbe::RangeFile::getIonID(), AtomProbe::RangeFile::getName(), AtomProbe::RANGE_MOLECULE::isOK, AtomProbe::RANGE_MOLECULE::name, and AtomProbe::AbundanceData::symbolIndex().
Referenced by checkMassRangingCorrectness().

◆ getRecordReadErrString()
|
inline |
Definition at line 46 of file dataFiles.h.
◆ GetReducedHullPts()
| unsigned int AtomProbe::GetReducedHullPts | ( | const std::vector< IonHit > & | points, |
| float | reductionDim, | ||
| unsigned int * | progress, | ||
| bool(*)(bool) | callback, | ||
| std::vector< IonHit > & | pointResult | ||
| ) |
Obtain a set of points that are a subset of points the convex hull that are at least "reductionDim" away from the hull itself.
Definition at line 289 of file convexHull.cpp.
References ASSERT, callback(), computeConvexHull(), distanceToFacet(), dotProduct(), freeConvexHull(), HULL_SURFREDUCE_NEGATIVE_SCALE_FACT, NUM_IONS, pyramidVol(), SCALE, and triIsDegenerate().
◆ getRotationMatrix()
| gsl_matrix* AtomProbe::getRotationMatrix | ( | const Point3D & | pt, |
| float | theta | ||
| ) |
Definition at line 246 of file rotations.cpp.
References ASSERT, computeRotationMatrix(), computeRotationMatrixWahba(), gsl_determinant(), AtomProbe::Point3D::normalise(), quat_rot(), AtomProbe::Point3D::sqrDist(), TEST, and AtomProbe::Point3D::transform3x3().
◆ gsl_determinant()
| float AtomProbe::gsl_determinant | ( | const gsl_matrix * | m | ) |
Definition at line 143 of file helpFuncs.cpp.
References ASSERT.
Referenced by computeRotationMatrix(), computeRotationMatrixWahba(), getRotationMatrix(), and AtomProbe::ComparePairFirstReverse::operator()().
◆ gsl_matrix_det()
| double AtomProbe::gsl_matrix_det | ( | gsl_matrix * | A, |
| bool | inPlace = false |
||
| ) |
Definition at line 37 of file rotations.cpp.
◆ gsl_matrix_mult()
| void AtomProbe::gsl_matrix_mult | ( | const gsl_matrix * | A, |
| const gsl_matrix * | B, | ||
| gsl_matrix *& | res, | ||
| bool | alloc = false |
||
| ) |
Definition at line 24 of file misc.cpp.
References ASSERT.
Referenced by computeRotationMatrixWahba(), and main().
◆ gsl_print_matrix()
| void AtomProbe::gsl_print_matrix | ( | const gsl_matrix * | m | ) |
Definition at line 122 of file helpFuncs.cpp.
Referenced by computeRangeAdjacency(), and AtomProbe::ComparePairFirstReverse::operator()().
◆ gsl_print_vector()
| void AtomProbe::gsl_print_vector | ( | const gsl_vector * | v | ) |
Definition at line 134 of file helpFuncs.cpp.
Referenced by AtomProbe::ComparePairFirstReverse::operator()().
◆ hexStrToUChar()
| void AtomProbe::hexStrToUChar | ( | const std::string & | s, |
| unsigned char & | c | ||
| ) |
◆ incrementCorrelationHist()
| bool AtomProbe::incrementCorrelationHist | ( | const std::vector< EPOS_ENTRY > & | eposIons, |
| std::vector< std::vector< unsigned int > > & | histogram, | ||
| float | stepMass, | ||
| float | endMass | ||
| ) |
Definition at line 29 of file histogram.cpp.
Referenced by accumulateCorrelationHistogram(), and correlationHistogram().
◆ intersect_RayTriangle()
| int AtomProbe::intersect_RayTriangle | ( | const Point3D & | rayStart, |
| const Point3D & | rayEnd, | ||
| Point3D * | tri, | ||
| Point3D & | I | ||
| ) |
Definition at line 280 of file mesh.cpp.
References AtomProbe::Point3D::crossProd(), AtomProbe::Point3D::dotProd(), AtomProbe::Point3D::normalise(), and AtomProbe::Point3D::sqrMag().
Referenced by AtomProbe::Mesh::pointsInside().
◆ isNotDirectory()
| bool AtomProbe::isNotDirectory | ( | const char * | filename | ) |
Definition at line 170 of file helpFuncs.cpp.
Referenced by AtomProbe::RangeFile::detectFileType(), loadTextData(), and AtomProbe::ComparePairFirstReverse::operator()().
◆ isValidElementSymbol()
| bool AtomProbe::isValidElementSymbol | ( | std::string & | symbol | ) |
Definition at line 86 of file ranges.cpp.
◆ kahansum()
| std::iterator_traits<T>::value_type AtomProbe::kahansum | ( | T | begin, |
| T | end, | ||
| T2 | other | ||
| ) |
Definition at line 49 of file confidence.cpp.
Referenced by cumTrapezoid().
◆ leastSquaresOverlapSolve()
| bool AtomProbe::leastSquaresOverlapSolve | ( | const AtomProbe::MultiRange & | rangeData, |
| const AtomProbe::AbundanceData & | abundance, | ||
| const std::vector< IonHit > & | hits, | ||
| float(*)(float, float) | backgroundEstimator, | ||
| std::vector< std::pair< unsigned int, float > > & | decomposedHits, | ||
| float | rangeTolerance = 0 |
||
| ) |
Solve overlaps using least squares for given multi-range.
Definition at line 47 of file deconvolution.cpp.
References AtomProbe::AbundanceData::abundanceBetweenLimits(), AtomProbe::MultiRange::addIon(), AtomProbe::MultiRange::addRange(), ASSERT, AtomProbe::SIMPLE_SPECIES::atomicNumber, AtomProbe::RGBf::blue, AtomProbe::SIMPLE_SPECIES::count, estimateRank(), AtomProbe::MultiRange::getIonName(), AtomProbe::MultiRange::getNumIons(), AtomProbe::MultiRange::getNumRanges(), AtomProbe::RandNumGen::getRng(), AtomProbe::RGBf::green, AtomProbe::AbundanceData::isotopes(), meanAndStdev(), AtomProbe::AbundanceData::open(), randGen, AtomProbe::RGBf::red, AtomProbe::MultiRange::setRangeGroups(), solveLeastSquares(), AtomProbe::MultiRange::splitOverlapping(), AtomProbe::AbundanceData::symbolIdxFromAtomicNumber(), AtomProbe::AbundanceData::symbolIndex(), and WARN.
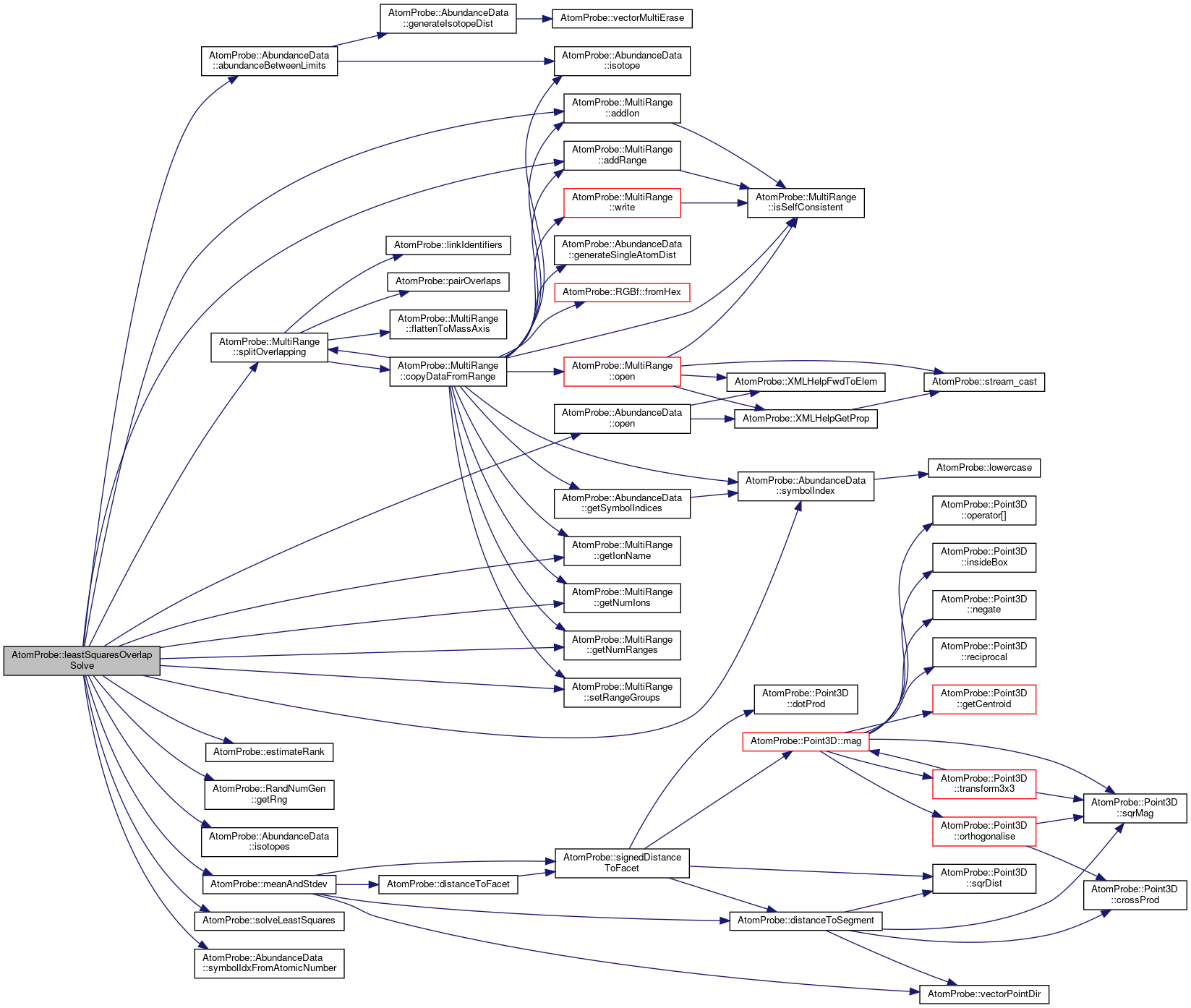
◆ likeLogGaussian()
| void AtomProbe::likeLogGaussian | ( | const std::vector< double > & | x, |
| double & | xp, | ||
| double & | sigma, | ||
| double & | lambda, | ||
| double & | amp, | ||
| double & | h, | ||
| std::vector< double > & | y | ||
| ) |
Generate a smoothed log-gaussian curve.
xp : positioning lambda :asymmetry h : FWHM (>0) amp : vertical rescale sigma : width
Definition at line 510 of file fitting.cpp.
References M_PI.
Referenced by fitLikeLogGaussian().
◆ linearHistogram()
| void AtomProbe::linearHistogram | ( | const std::vector< T > & | data, |
| T | start, | ||
| T | end, | ||
| T | step, | ||
| std::vector< T > & | histVals | ||
| ) |
Make a linearly spaced histogram with the given spacings Takes a vector of floats as input,
- Parameters
-
data
Definition at line 75 of file histogram.h.
References ASSERT.
Referenced by main().
◆ linkIdentifiers()
| void AtomProbe::linkIdentifiers | ( | vector< T > & | link | ) |
Definition at line 88 of file multiRange.cpp.
Referenced by AtomProbe::MultiRange::splitOverlapping().
◆ loadATOFile()
| unsigned int AtomProbe::loadATOFile | ( | const char * | fileName, |
| std::vector< ATO_ENTRY > & | ions, | ||
| unsigned int | forceEndian = 0 |
||
| ) |
Load a LAWATAP "ATO" file.
ATO files contain a mix of experiment and analysis data, including voltage, positioning and reconstructed data. There are specific algorithm generated data (eg clustering) which are not loaded in structures in this library to reduce memory consumption. Returns 0 on success, nonzero on error
- Filename : the name of the data file to read
- ions : the returned entries from the file
- forceEndian : if nonzero, this wil force the use of either little-endian reading (1), or big-endian(2). If zero, the routine will try to automatically determine endianness from the file contents.
Returned code if nonzero can be converted to human readable form using ATO_ERR_STRINGS. Note if calling from outside C++, use getAtoErrString(...) to return the error string
Definition at line 1212 of file dataFiles.cpp.
References ASSERT, ATO_EMPTY_FAIL, ATO_MEM_ERR, ATO_OPEN_FAIL, ATO_SIZE_ERR, ATO_VERSIONCHECK_ERR, floatSwapBytes(), AtomProbe::RandNumGen::getRng(), randGen, and randomIndices().
◆ loadEposFile()
| size_t AtomProbe::loadEposFile | ( | std::vector< EPOS_ENTRY > & | outData, |
| const char * | filename | ||
| ) |
Load an entire "EPOS" File.
Epos files are very large and unwieldy. Ensure that you really want to do this, as it is possible you will run out of memory. Consider using the "chunkLoadEposFile" to process your data if possible. The file contains x,y,z,m-to-c,tof(uncorrected?). The return value is nonzero on error, and is part of the RECORDREAD_ERR enum
Definition at line 756 of file dataFiles.cpp.
References EPOS_RECORD_SIZE, fixedRecordReader(), and readEposRecord().
Referenced by getAtoErrString(), and main().

◆ loadPosFile() [1/2]
| unsigned int AtomProbe::loadPosFile | ( | std::vector< IonHit > & | posIons, |
| const char * | posFile | ||
| ) |
Load a pos file directly into a single ion list.
Load a pos file from a file. Returns nonzero on exit.
Pos files are fixed record size files, with data stored as 4byte big endian floating point. (IEEE 754?). Data is stored as x,y,z,mass/charge.
Currently loads entire file in one gulp. This is bad from the point of view of gigabyte size pos files generated by laser atom probes.. Returns 0 on ok, nonzero on success.
arguments : posfile | path to pos-formatted file posIons | will return any ions that are loaded from the dataset. Behaviour for loading into * a non-empty vector is undefined Return code can be decoded with getPosFileErrString
Definition at line 256 of file dataFiles.cpp.
References ASSERT, AtomProbe::IonHit::hasNaN(), AtomProbe::IONHIT::massToCharge, AtomProbe::IONHIT::pos, POS_ALLOC_FAIL, POS_NAN_LOAD_ERROR, POS_OPEN_FAIL, POS_READ_FAIL, POS_SIZE_EMPTY_ERR, POS_SIZE_MODULUS_ERR, AtomProbe::IonHit::setMassToCharge(), and AtomProbe::IonHit::setPos().
Referenced by getAtoErrString(), loadPosFile(), and main().
◆ loadPosFile() [2/2]
| unsigned int AtomProbe::loadPosFile | ( | std::vector< IonHit > & | posIons, |
| const char * | posFile, | ||
| unsigned int | nSamplesMax | ||
| ) |
As per loadPosFile, but with an additional setting for the maximum number of ions to load.
Ions will be randomly sampled to form nSamples. This will be likely much slower than loading a full file, depending on the size of nSamples. The samples are not guaranteed to be the same for each load. Speed will be nlogn in nSamplesMax, o(1) in posIons.
Definition at line 364 of file dataFiles.cpp.
References AtomProbe::RandNumGen::getRng(), loadPosFile(), AtomProbe::IONHIT::massToCharge, AtomProbe::IONHIT::pos, POS_ALLOC_FAIL, POS_NAN_LOAD_ERROR, POS_OPEN_FAIL, POS_READ_FAIL, POS_SIZE_EMPTY_ERR, POS_SIZE_MODULUS_ERR, randGen, and randomIndices().
◆ loadTapsimBinFile()
| unsigned int AtomProbe::loadTapsimBinFile | ( | vector< IonHit > & | posIons, |
| const char * | posfile | ||
| ) |
Definition at line 538 of file dataFiles.cpp.
References AtomProbe::IonHit::setMassToCharge(), AtomProbe::IonHit::setPos(), splitStrsRef(), stream_cast(), TAPSIM_FILE_FORMAT_FAIL, and TAPSIM_OPEN_FAIL.
◆ loadTextData()
| unsigned int AtomProbe::loadTextData | ( | const char * | cpFilename, |
| std::vector< std::vector< float > > & | dataVec, | ||
| std::vector< std::string > & | headerVec, | ||
| const char * | delim = "\t,", |
||
| bool | allowNan = true, |
||
| bool | allowConvFails = false, |
||
| unsigned int | headerCount = -1 |
||
| ) |
Load a CSV, TSV or similar text file. Assumes "C" Locale for input (ie "." as decimal separator).
Maximum line length is 4K. Data will be given out as a vector v[Column][line] in the input text file. this allows for ready column extraction.
headerVec - strings used to represent columns allowConvFails - replace strings that cannot be converted to float with NaN allowNan - return error if the input file contains nan headerCount - the maximum number of lines to consider before declaring that line as the header. if -1, this parameter has no effect
Note the function transposeVector may be of interest
Definition at line 1504 of file dataFiles.cpp.
References isNotDirectory(), splitStrsRef(), stream_cast(), strhas(), stripZeroEntries(), TEXT_ERR_FILE_FORMAT, TEXT_ERR_FILE_NUM_FIELDS, and TEXT_ERR_FILE_OPEN.
Referenced by getAtoErrString(), and main().
◆ lowercase()
| std::string AtomProbe::lowercase | ( | std::string | s | ) |
Return a lowercase version for a given string.
Definition at line 176 of file stringFuncs.cpp.
Referenced by AtomProbe::RangeFile::detectFileType(), main(), parseCompositionData(), AtomProbe::RangeFile::rangeTypeString(), strAppend(), and AtomProbe::AbundanceData::symbolIndex().
◆ lsq()
| double AtomProbe::lsq | ( | const std::vector< double > & | y, |
| const std::vector< double > & | yFit | ||
| ) |
Definition at line 27 of file fitting.cpp.
References voigtProfile(), AtomProbe::DATA_FIT::x, and AtomProbe::DATA_FIT::y.
Referenced by fitDoniachSunjic(), fitExpNorm(), fitLikeLogGaussian(), fitVoigtFunc(), and main().
◆ makeHistogram()
| void AtomProbe::makeHistogram | ( | const vector< float > & | data, |
| float | start, | ||
| float | end, | ||
| float | step, | ||
| vector< float > & | histVals | ||
| ) |
Definition at line 126 of file processing.cpp.
References ASSERT.
Referenced by doFitBackground(), and findPeaks().
◆ marchingCubes()
| void AtomProbe::marchingCubes | ( | const Voxels< float > & | v, |
| float | isoValue, | ||
| std::vector< TriangleWithVertexNorm > & | tVec | ||
| ) |
Definition at line 494 of file isoSurface.cpp.
References ASSERT, AtomProbe::BoundCube::containsPt(), AtomProbe::Voxels< T >::deprecatedGetEdgeUniqueIndex(), AtomProbe::Voxels< T >::fill(), AtomProbe::Voxels< T >::getBounds(), AtomProbe::Voxels< T >::getData(), AtomProbe::Voxels< T >::getEdgeEndApproxVals(), AtomProbe::Voxels< T >::getEdgeEnds(), AtomProbe::Voxels< T >::getMaxBounds(), AtomProbe::Voxels< T >::getMinBounds(), AtomProbe::Voxels< T >::getPitch(), AtomProbe::Voxels< T >::getSize(), AtomProbe::TriangleWithVertexNorm::isDegenerate(), AtomProbe::TriangleWithVertexNorm::normal, AtomProbe::TriangleWithVertexNorm::p, AtomProbe::TriangleWithIndexedVertices::p, AtomProbe::TRIANGLE::p, AtomProbe::TRIANGLE::physGroup, removeElements(), AtomProbe::Voxels< T >::resize(), AtomProbe::TriangleWithVertexNorm::safeComputeACWNormal(), AtomProbe::BoundCube::setBounds(), AtomProbe::Voxels< T >::setData(), and TEST.
Referenced by main().
◆ matchComposedName()
| bool AtomProbe::matchComposedName | ( | const std::map< string, size_t > & | composedNames, |
| const vector< pair< string, size_t > > & | namesToFind, | ||
| size_t & | matchOffset | ||
| ) |
Definition at line 297 of file ranges.cpp.
References AtomProbe::RangeFile::decomposeIonNames().
Referenced by AtomProbe::RangeFile::open().

◆ maxExplainedFraction() [1/2]
| std::vector<float> AtomProbe::maxExplainedFraction | ( | const std::vector< std::pair< float, float > > & | massData, |
| float | peakMass, | ||
| float | massWidth, | ||
| const std::vector< std::vector< AtomProbe::ISOTOPE_ENTRY > > & | solutions, | ||
| const AtomProbe::AbundanceData & | massTable, | ||
| float | massDistTol, | ||
| unsigned int | solutionCharge | ||
| ) |
Compute the fraction of the data that has been explained, using the natural abundance information, and intensity data.
Specifically, this returns the maximum possible fraction of this peak that is explained by the isotopic fingerprint for a given solution set, and the count data given. This is returned as a vector with a value for each proposed solution Note the solutions do not need to sum to 1
massData - list of location and intensities for peaks in dataset peakMass - peak we wish to query. Must be present (within tolerance) in peak list (massData) massWidth - tolerance width (+-) when locating peaks solutions - list of candidate solutions we want to find the explained fraction for massDistTol - tolerance to use when checking for peaks in mass distribution solutionCharge - charge state to apply to isotope distribuion
Referenced by maxExplainedFraction().
◆ maxExplainedFraction() [2/2]
| vector<float> AtomProbe::maxExplainedFraction | ( | const vector< pair< float, float > > & | intensityData, |
| float | peakMass, | ||
| float | massWidth, | ||
| const vector< vector< ISOTOPE_ENTRY > > & | solutions, | ||
| const AbundanceData & | massTable, | ||
| float | massDistTol, | ||
| unsigned int | solutionCharge | ||
| ) |
Definition at line 221 of file filter.cpp.
References ASSERT, buildFrequencyTable(), countIntensityEvents(), AtomProbe::AbundanceData::generateGroupedIsotopeDist(), AtomProbe::AbundanceData::getMajorIsotopeFromElemIdx(), AtomProbe::AbundanceData::isotope(), AtomProbe::ISOTOPE_ENTRY::mass, maxExplainedFraction(), AtomProbe::AbundanceData::open(), AtomProbe::AbundanceData::symbolIndex(), and TEST.
◆ meanAndStdev()
| void AtomProbe::meanAndStdev | ( | const std::vector< T > & | f, |
| float & | meanVal, | ||
| float & | stdevVal, | ||
| bool | normalCorrection = true |
||
| ) |
Definition at line 76 of file misc.h.
References distanceToFacet(), distanceToSegment(), signedDistanceToFacet(), and vectorPointDir().
Referenced by andersonDarlingStatistic(), and leastSquaresOverlapSolve().
◆ myStrDup()
| char* AtomProbe::myStrDup | ( | const char * | s | ) |
Definition at line 39 of file helpFuncs.cpp.
Referenced by pushLocale().
◆ nullBackground()
| float AtomProbe::nullBackground | ( | float | rangeStart, |
| float | rangeEnd | ||
| ) |
Definition at line 41 of file deconvolution.cpp.
◆ nullifyMarker()
| void AtomProbe::nullifyMarker | ( | char * | buffer, |
| char | marker | ||
| ) |
Definition at line 55 of file stringFuncs.cpp.
Referenced by AtomProbe::RangeFile::rangeTypeString(), and stlStrToStlWStr().
◆ numericalEstimateGaussRatioConf()
| bool AtomProbe::numericalEstimateGaussRatioConf | ( | float | mu1, |
| float | mu2, | ||
| float | var1, | ||
| float | var2, | ||
| float | alpha, | ||
| unsigned int | nTrials, | ||
| gsl_rng * | r, | ||
| float & | lBound, | ||
| float & | uBound | ||
| ) |
Brute-force guassian ratio confidence estimator.
Performs monte-carlo trials to determine the distribution of Z=X/Y. where X~Gaussian(L1), Y~max(Gaussian(L2),1)
Biased, as it enforces 1, in the denominator of L1/L2 (ie L2=0 has p=0) returns false if either bound could not be computed
- mu1 : Mean 1 (must be +ve)
- mu2 : Mean 2 (must be +ve)
- var1 : Variance 1 (must be +ve)
- var2 : Variance 2 (must be +ve)
- alpha : confidence interval in distribution
- nTrials : number of computations of Z.
- lBound : returned lower bound at confidence alpha
- uBound : returned upper bound at confidence alpha
Note that the estimator seems to biased to the number of trials, and needs to be improved.
Definition at line 275 of file confidence.cpp.
References ASSERT.
Referenced by poissonConfidenceObservation().
◆ numericalEstimatePoissRatioConf()
| bool AtomProbe::numericalEstimatePoissRatioConf | ( | float | lambda1, |
| float | lambda2, | ||
| float | alpha, | ||
| unsigned int | nTrials, | ||
| gsl_rng * | r, | ||
| float & | lBound, | ||
| float & | uBound | ||
| ) |
Brute-force poisson ratio confidence estimator. Returns the confidence interval in the estimate of the mean, at the given rates.
Performs monte-carlo trials to determine the distribution of Z=X/Y. where X~Poisson(L1), Y~Poisson(L2) Biased, as it enforces 1, in the denominator of L1/L2 (ie L2=0 has p=0) returns false if either bound could not be computed
- lambda1 : Poisson rate 1 (must be +ve)
- lambda2 : Poisson rate 2 (must be +ve)
- alpha : confidence interval in distribution
- nTrials : number of computations of Z.
- lBound : returned lower bound at confidence alpha. This is interpolated, so is a real number
- uBound : returned upper bound at confidence alpha. This is interpolated, so is a real number.
Note that the estimator seems to slightly biased to the number of trials, and needs to be improved.
Definition at line 68 of file confidence.cpp.
References ASSERT.
Referenced by main(), and poissonConfidenceObservation().
◆ numericalEstimateSkellamConf()
| bool AtomProbe::numericalEstimateSkellamConf | ( | float | lambda1, |
| float | lambda2, | ||
| float | alpha, | ||
| unsigned int | nTrials, | ||
| gsl_rng * | r, | ||
| float & | lBound, | ||
| float & | uBound | ||
| ) |
Brute-force the confidence bounds of a skellam distribution.
Performs monte-carlo trials to determine Z = X - Y, where X, Y are poisson (Lambda1, Lambda2).
- Lambda1 : poisson rate for X
- Lambda2: poisson rate for Y
- alpha : confidence bound
- nTrials : number of computations of Z.
- lBound : returned lower bound at confidence alpha. This is interpolated, so is a real number
- uBound : returned upper bound at confidence alpha. This is interpolated, so is a real number
Definition at line 173 of file confidence.cpp.
References ASSERT.
Referenced by poissonConfidenceObservation().
◆ onlyDir()
| std::string AtomProbe::onlyDir | ( | const std::string & | path | ) |
Return only the directory name component of the full path.
Definition at line 44 of file stringFuncs.cpp.
Referenced by strAppend().
◆ onlyFilename()
| std::string AtomProbe::onlyFilename | ( | const std::string & | path | ) |
Return only the filename component.
Definition at line 33 of file stringFuncs.cpp.
Referenced by strAppend().
◆ operator<()
| bool AtomProbe::operator< | ( | const SIMPLE_SPECIES & | a, |
| const SIMPLE_SPECIES & | b | ||
| ) |
Definition at line 117 of file multiRange.cpp.
References AtomProbe::SIMPLE_SPECIES::atomicNumber, and AtomProbe::SIMPLE_SPECIES::count.
◆ operator<<() [1/3]
| std::ostream& AtomProbe::operator<< | ( | std::ostream & | stream, |
| const IonHit & | ion | ||
| ) |
Definition at line 181 of file ionhit.cpp.
References AtomProbe::IonHit::getMassToCharge(), and AtomProbe::IonHit::getPos().
◆ operator<<() [2/3]
| std::ostream& AtomProbe::operator<< | ( | std::ostream & | stream, |
| const Point3D & | pt | ||
| ) |
Definition at line 417 of file point3D.cpp.
◆ operator<<() [3/3]
| std::ostream& AtomProbe::operator<< | ( | std::ostream & | stream, |
| const BoundCube & | b | ||
| ) |
Definition at line 563 of file boundcube.cpp.
◆ pairContains()
| bool AtomProbe::pairContains | ( | const pair< float, float > & | p, |
| float | m | ||
| ) |
Definition at line 46 of file rangeCheck.cpp.
References ASSERT.
Referenced by checkMassRangingCorrectness().
◆ pairOverlaps()
| bool AtomProbe::pairOverlaps | ( | float | aStart, |
| float | aEnd, | ||
| float | bStart, | ||
| float | bEnd | ||
| ) |
Definition at line 122 of file multiRange.cpp.
References ASSERT.
Referenced by AtomProbe::MultiRange::splitOverlapping().
◆ parseColString()
| bool AtomProbe::parseColString | ( | const std::string & | str, |
| unsigned char & | r, | ||
| unsigned char & | g, | ||
| unsigned char & | b, | ||
| unsigned char & | a | ||
| ) |
Parse a colour string containing rgb[a]; hex for with leading #.
Definition at line 269 of file stringFuncs.cpp.
References hexStrToUChar().
Referenced by AtomProbe::RGBf::fromHex(), AtomProbe::MultiRange::open(), AtomProbe::RangeFile::rangeTypeString(), and strAppend().

◆ parseCompositionData()
| unsigned int AtomProbe::parseCompositionData | ( | const std::vector< std::string > & | s, |
| const AbundanceData & | atomTable, | ||
| std::map< unsigned int, double > & | atomData | ||
| ) |
Convert atomic string label data, in the form "Fe 0.81" to fraction map.
The input type (mol/mass) is not relevant for this function, but should be in fraction the label "bal" can be used in place of a composition value exactly once to provide "balance" mass to sum to 1 returns 0 on success, nonzero on error (see COMPPARSE enum)
Definition at line 100 of file massconvert.cpp.
References COMPPARSE_BAD_BAL_USAGE, COMPPARSE_BAD_COMP_VALUE, COMPPARSE_BAD_INPUT_FORMAT, COMPPARSE_BAD_SYMBOL, COMPPARSE_BAD_TOTAL_COMP, COMPPARSE_DUPLICATE_SYMBOL, convertMassToMol(), convertMolToMass(), lowercase(), AtomProbe::AbundanceData::open(), splitStrsRef(), stream_cast(), stripWhite(), stripZeroEntries(), AtomProbe::AbundanceData::symbolIndex(), and TEST.
◆ poissonConfidenceObservation()
| void AtomProbe::poissonConfidenceObservation | ( | float | counts, |
| float | alpha, | ||
| float & | lBound, | ||
| float & | uBound | ||
| ) |
Obtain poisson confidence limits for the mean of a poisson distribution.
Definition at line 508 of file confidence.cpp.
References ASSERT, AtomProbe::RandNumGen::getRng(), numericalEstimateGaussRatioConf(), numericalEstimatePoissRatioConf(), numericalEstimateSkellamConf(), randGen, TEST, and zechConfidenceLimits().
◆ popLocale()
| void AtomProbe::popLocale | ( | ) |
Definition at line 96 of file helpFuncs.cpp.
Referenced by AtomProbe::RangeFile::openFormat().
◆ pushLocale()
| void AtomProbe::pushLocale | ( | const char * | newLocale, |
| int | type | ||
| ) |
Definition at line 62 of file helpFuncs.cpp.
References ASSERT, and myStrDup().
Referenced by AtomProbe::RangeFile::openFormat().

◆ pyramidVol()
Definition at line 290 of file misc.cpp.
References ASSERT, det3by3(), and estimateRank().
Referenced by dotProduct(), and GetReducedHullPts().
◆ quat_get_rot_quat()
| void AtomProbe::quat_get_rot_quat | ( | const Point3f * | rotVec, |
| float | angle, | ||
| Quaternion * | rotQuat | ||
| ) |
Compute the quaternion for specified rotation.
angle is in radians. pass to quat_rot_apply_quat to compute rotation
Definition at line 184 of file quat.cpp.
References AtomProbe::Quaternion::a, ASSERT, AtomProbe::Quaternion::b, AtomProbe::Quaternion::c, AtomProbe::Quaternion::d, AtomProbe::Point3f::fx, AtomProbe::Point3f::fy, and AtomProbe::Point3f::fz.
Referenced by AtomProbe::Mesh::rotate().
◆ quat_invert()
| void AtomProbe::quat_invert | ( | Quaternion * | quat | ) |
Definition at line 219 of file quat.cpp.
References AtomProbe::Quaternion::b, AtomProbe::Quaternion::c, and AtomProbe::Quaternion::d.
◆ quat_mult_no_second_a()
| void AtomProbe::quat_mult_no_second_a | ( | Quaternion * | result, |
| const Quaternion * | q1, | ||
| const Quaternion * | q2 | ||
| ) |
Definition at line 39 of file quat.cpp.
References AtomProbe::Quaternion::a, AtomProbe::Quaternion::b, AtomProbe::Quaternion::c, and AtomProbe::Quaternion::d.
Referenced by quat_rot(), quat_rot_apply_quat(), and quat_rot_array().
◆ quat_pointmult()
| void AtomProbe::quat_pointmult | ( | Point3f * | result, |
| const Quaternion * | q1, | ||
| const Quaternion * | q2 | ||
| ) |
Definition at line 50 of file quat.cpp.
References AtomProbe::Quaternion::a, AtomProbe::Quaternion::b, AtomProbe::Quaternion::c, AtomProbe::Quaternion::d, AtomProbe::Point3f::fx, AtomProbe::Point3f::fy, and AtomProbe::Point3f::fz.
Referenced by quat_rot(), quat_rot_apply_quat(), and quat_rot_array().
◆ quat_rot() [1/2]
Rotate a point around a given rotation axis by a specified angle.
This is not efficient for large numbers of points, as this will recompute the quaternion each time
Definition at line 59 of file quat.cpp.
References AtomProbe::Point3f::fx, AtomProbe::Point3f::fy, and AtomProbe::Point3f::fz.
Referenced by getRotationMatrix().
◆ quat_rot() [2/2]
Rotate a point around a given vector, with specified angle.
This is not efficient for large numbers of points, as this will recompute the quaternion each time. angle (radians) is anti-clockwise as viewed from positive side of vector (right-handed)
Definition at line 77 of file quat.cpp.
References AtomProbe::Quaternion::a, ASSERT, AtomProbe::Quaternion::b, AtomProbe::Quaternion::c, AtomProbe::Quaternion::d, AtomProbe::Point3f::fx, AtomProbe::Point3f::fy, AtomProbe::Point3f::fz, quat_mult_no_second_a(), and quat_pointmult().
◆ quat_rot_apply_quat()
| void AtomProbe::quat_rot_apply_quat | ( | Point3f * | point, |
| const Quaternion * | rotQuat | ||
| ) |
Use previously generated quats from quat_get_rot_quats to rotate a point.
Rotation is in-place, on single point
Definition at line 206 of file quat.cpp.
References AtomProbe::Quaternion::b, AtomProbe::Quaternion::c, AtomProbe::Quaternion::d, AtomProbe::Point3f::fx, AtomProbe::Point3f::fy, AtomProbe::Point3f::fz, quat_mult_no_second_a(), and quat_pointmult().
Referenced by AtomProbe::Mesh::rotate().
◆ quat_rot_array() [1/2]
| void AtomProbe::quat_rot_array | ( | Point3f * | point, |
| unsigned int | n, | ||
| const Point3f * | rotVec, | ||
| float | angle | ||
| ) |
Rotate each point in array of size n around a given vector, with specified angle.
rotVec is the rotation, angle is the anti-clockwise angle as viewed from positive side of vector ( right-handed) rotation of points is in-place
Definition at line 139 of file quat.cpp.
References AtomProbe::Quaternion::a, ASSERT, AtomProbe::Quaternion::b, AtomProbe::Quaternion::c, AtomProbe::Quaternion::d, AtomProbe::Point3f::fx, AtomProbe::Point3f::fy, AtomProbe::Point3f::fz, quat_mult_no_second_a(), and quat_pointmult().
Referenced by quat_rot_array().
◆ quat_rot_array() [2/2]
| void AtomProbe::quat_rot_array | ( | Point3D * | point, |
| unsigned int | n, | ||
| const Point3f * | rotVec, | ||
| float | angle | ||
| ) |
Rotate each point in array of size n around a given vector, with specified angle.
rotVec is the rotation, angle is the anti-clockwise angle as viewed from positive side of vector ( right-handed). rotation of points is in-place
Definition at line 115 of file quat.cpp.
References AtomProbe::Point3f::fx, AtomProbe::Point3f::fy, AtomProbe::Point3f::fz, and quat_rot_array().
◆ randomIndices()
| void AtomProbe::randomIndices | ( | std::vector< T > & | res, |
| size_t | num, | ||
| size_t | nMax, | ||
| gsl_rng * | rng | ||
| ) |
Definition at line 113 of file sampling.h.
Referenced by loadATOFile(), and loadPosFile().
◆ randomSelect()
| size_t AtomProbe::randomSelect | ( | std::vector< T > & | result, |
| const std::vector< T > & | source, | ||
| size_t | num, | ||
| gsl_rng * | rng | ||
| ) |
Definition at line 40 of file sampling.h.
References AtomProbe::LinearFeedbackShiftReg::clock(), AtomProbe::LinearFeedbackShiftReg::setMaskPeriod(), and AtomProbe::LinearFeedbackShiftReg::setState().
Referenced by sampleIons().
◆ rangeOverlaps()
| bool AtomProbe::rangeOverlaps | ( | const pair< float, float > & | r1, |
| const pair< float, float > & | r2 | ||
| ) |
Definition at line 150 of file componentAnalysis.cpp.
Referenced by computeRangeAdjacency().
◆ rangeToStr()
| string AtomProbe::rangeToStr | ( | const pair< float, float > & | rng | ) |
Definition at line 111 of file ranges.cpp.
References stream_cast().
Referenced by AtomProbe::RangeFile::isSelfConsistent().

◆ readEposRecord()
| bool AtomProbe::readEposRecord | ( | const char * | src, |
| const char * | dest | ||
| ) |
Definition at line 711 of file dataFiles.cpp.
References AtomProbe::EPOS_ENTRY::deltaPulse, floatSwapBytes(), AtomProbe::EPOS_ENTRY::hitMultiplicity, int4SwapBytes(), AtomProbe::EPOS_ENTRY::massToCharge, AtomProbe::EPOS_ENTRY::timeOfFlight, AtomProbe::EPOS_ENTRY::voltDC, AtomProbe::EPOS_ENTRY::voltPulse, AtomProbe::EPOS_ENTRY::x, AtomProbe::EPOS_ENTRY::xDetector, AtomProbe::EPOS_ENTRY::y, AtomProbe::EPOS_ENTRY::yDetector, and AtomProbe::EPOS_ENTRY::z.
Referenced by chunkLoadEposFile(), and loadEposFile().
◆ readPosapOps()
| unsigned int AtomProbe::readPosapOps | ( | const char * | file, |
| THREEDAP_EXPERIMENT & | data, | ||
| unsigned int & | badLine, | ||
| unsigned int & | progress, | ||
| std::atomic< bool > & | wantAbort, | ||
| unsigned int | nDelayLines = 2, |
||
| bool | strictMode = false |
||
| ) |
Function to read POSAP "OPS" files.
OPS files contain data on voltage, positioned hits and time of flights as well as some instrument parameters. This format is not standardised, so variants may exist
- badLine returns the line on which an error was encountered.
- progress gives a 0-100% progress value (counted in file size). Can be used to multithread monitor progress
- nDelayLines - The number of delay lines in the system. Usually 2, possibly 3.
- strictMode - enable to throw errors in cases where files appear to be badly formatted TODO: Can we work out how to obtain nDelayLines from parsing the file?
Definition at line 805 of file dataFiles.cpp.
References AtomProbe::THREEDAP_DATA::alpha, ARRAYSIZE, ASSERT, AtomProbe::VOLTAGE_DATA::beta, AtomProbe::THREEDAP_DATA::beta, AtomProbe::THREEDAP_DATA::detectorChannels, AtomProbe::THREEDAP_DATA::detectorRadius, AtomProbe::THREEDAP_EXPERIMENT::eventData, AtomProbe::THREEDAP_EXPERIMENT::eventPulseNumber, AtomProbe::THREEDAP_DATA::flightPath, getFilesize(), AtomProbe::VOLTAGE_DATA::nextHitGroupOffset, OPSREADER_ABORT_ERR, OPSREADER_CHANNELS_DATA_ERR, OPSREADER_ENUM_END, OPSREADER_FORMAT_CHANNELS_ERR, OPSREADER_FORMAT_CLINE_ERR, OPSREADER_FORMAT_DETECTORLINE_ERR, OPSREADER_FORMAT_DOUBLEDASH, OPSREADER_FORMAT_DUPLICATE_DETECTORSIZE, OPSREADER_FORMAT_DUPLICATE_SYSDATA, OPSREADER_FORMAT_LINE_DASH_ERR, OPSREADER_FORMAT_LINE_VOLTAGE_DATA_ERR, OPSREADER_FORMAT_LINE_VOLTAGE_DATACOUNT_ERR, OPSREADER_FORMAT_LINE_VOLTAGE_NOBETA, OPSREADER_FORMAT_LINETYPE_ERR, OPSREADER_FORMAT_SLINE_EVENTCOUNT_ERR, OPSREADER_FORMAT_SLINE_EVENTDATA_ERR, OPSREADER_FORMAT_SLINE_FORMAT_ERR, OPSREADER_FORMAT_SLINE_PREFIX_ERR, OPSREADER_FORMAT_TRAILING_DASH_ERR, OPSREADER_OPEN_ERR, OPSREADER_READ_ERR, AtomProbe::THREEDAP_EXPERIMENT::params, AtomProbe::VOLTAGE_DATA::pulseVolt, splitStrsRef(), stream_cast(), stripWhite(), stripZeroEntries(), AtomProbe::SINGLE_HIT::tof, AtomProbe::THREEDAP_DATA::tZero, AtomProbe::VOLTAGE_DATA::voltage, and AtomProbe::THREEDAP_EXPERIMENT::voltageData.
◆ reconstructTest()
| bool AtomProbe::reconstructTest | ( | ) |
Referenced by AtomProbe::ReconstructionSphereOnCone::reconstruct(), and runTests().
◆ removeElements()
| void AtomProbe::removeElements | ( | const std::vector< size_t > & | elems, |
| std::vector< T > & | vec | ||
| ) |
◆ rotateMatch()
| bool AtomProbe::rotateMatch | ( | const unsigned int * | a, |
| const unsigned int * | b, | ||
| size_t | n, | ||
| bool | directionForwards = true |
||
| ) |
Definition at line 100 of file mesh.cpp.
Referenced by AtomProbe::TRIANGLE::edgesMismatch().
◆ sampleIons()
| void AtomProbe::sampleIons | ( | const std::vector< IonHit > & | ions, |
| float | sampleFactor, | ||
| std::vector< IonHit > & | sampled, | ||
| bool | strongRandom = true |
||
| ) |
Definition at line 30 of file sampling.cpp.
References AtomProbe::RandNumGen::getRng(), randGen, randomSelect(), and TEST.
Referenced by main().
◆ savePosFile() [1/2]
| unsigned int AtomProbe::savePosFile | ( | const std::vector< Point3D > & | points, |
| float | mass, | ||
| const char * | name, | ||
| bool | append = false |
||
| ) |
Save a vector of Point3Ds into a pos file, using a fixed mass, return nonzero on error.
Create an pos file from a vector of points, and a given mass.
A nonzero return code can be decoded with getPosFileErrString, which will provide a human readable error message
- if append is true, the the file will be appended to.
Definition at line 499 of file dataFiles.cpp.
References floatSwapBytes().
Referenced by getAtoErrString(), main(), and AtomProbe::ReconstructionSphereOnCone::reconstruct().
◆ savePosFile() [2/2]
| unsigned int AtomProbe::savePosFile | ( | const std::vector< IonHit > & | data, |
| const char * | name, | ||
| bool | append = false |
||
| ) |
Save a vector of IonHits into a "pos" file, return nonzero on error.
Create an pos file from a vector of IonHits.
Definition at line 463 of file dataFiles.cpp.
References floatSwapBytes().
◆ saveTapsimBin() [1/2]
| unsigned int AtomProbe::saveTapsimBin | ( | const std::vector< IonHit > & | posIons, |
| std::ostream & | f | ||
| ) |
Write a tapsim file from a vector of IonHits.
Note: there is some data loss here. Tapsim data basically writes a short header and then the ion data is little endian stored, not big-endian. Finally, m/c data is written as short, hence data loss
Definition at line 667 of file dataFiles.cpp.
References ASSERT, floatSwapBytes(), and stream_cast().
Referenced by getAtoErrString(), and saveTapsimBin().
◆ saveTapsimBin() [2/2]
| unsigned int AtomProbe::saveTapsimBin | ( | const std::vector< IonHit > & | posIons, |
| const char * | filename | ||
| ) |
Write a tapsim file from a vector of ionHits.
Convenience Wrapper for other saveTapsimBin(...) function
Definition at line 703 of file dataFiles.cpp.
References saveTapsimBin().
◆ selectElements()
| void AtomProbe::selectElements | ( | const std::vector< T > & | in, |
| const std::vector< unsigned int > & | indices, | ||
| std::vector< T > & | out | ||
| ) |
Obtain the elements at positions indicies in the input vector, copy to output.
Definition at line 67 of file misc.h.
Referenced by findPeaks().
◆ setHardAssert()
| void AtomProbe::setHardAssert | ( | bool | enabled | ) |
Do assertions cause a straight up crash (enabled), or write a mesage to stderr (disabled)? By default, hard crash.
Definition at line 30 of file aptAssert.cpp.
◆ signedDistanceToFacet()
| float AtomProbe::signedDistanceToFacet | ( | const Point3D & | fA, |
| const Point3D & | fB, | ||
| const Point3D & | fC, | ||
| const Point3D & | normal, | ||
| const Point3D & | p | ||
| ) |
Find the distance between a point, and a triangular facet – may be positive or negative.
Definition at line 172 of file misc.cpp.
References ASSERT, distanceToSegment(), AtomProbe::Point3D::dotProd(), AtomProbe::Point3D::mag(), and AtomProbe::Point3D::sqrDist().
Referenced by distanceToFacet(), AtomProbe::Mesh::getNearestTri(), and meanAndStdev().
◆ signVal()
| float AtomProbe::signVal | ( | unsigned int | val | ) |
Definition at line 147 of file mesh.cpp.
Referenced by Determinant().
◆ solveLeastSquares()
| bool AtomProbe::solveLeastSquares | ( | gsl_matrix * | m, |
| gsl_vector * | b, | ||
| gsl_vector *& | x | ||
| ) |
Use an SVD based least-squares solver to solve Mx=b (for x).
Definition at line 94 of file misc.cpp.
Referenced by leastSquaresOverlapSolve().
◆ SphericProjectionEqn()
| double AtomProbe::SphericProjectionEqn | ( | double | eta, |
| void * | p | ||
| ) |
Definition at line 42 of file projection.cpp.
References AtomProbe::SphericProjectionParams::focusDist, and AtomProbe::SphericProjectionParams::theta.
Referenced by AtomProbe::StereographicProjection::thetaToEta(), and AtomProbe::ModifiedFocusSphericProjection::thetaToEta().
◆ splitStrsRef() [1/2]
| void AtomProbe::splitStrsRef | ( | const char * | cpStr, |
| const char | delim, | ||
| std::vector< std::string > & | v | ||
| ) |
Split string references using a single delimiter.
Definition at line 187 of file stringFuncs.cpp.
Referenced by AtomProbe::RangeFile::detectFileType(), AtomProbe::Mesh::loadGmshMesh(), loadTapsimBinFile(), loadTextData(), main(), AtomProbe::Point3D::parse(), parseCompositionData(), AtomProbe::RangeFile::rangeTypeString(), readPosapOps(), and strAppend().
◆ splitStrsRef() [2/2]
| void AtomProbe::splitStrsRef | ( | const char * | cpStr, |
| const char * | delim, | ||
| std::vector< std::string > & | v | ||
| ) |
Split string references using any of a given string of delimiters.
Definition at line 221 of file stringFuncs.cpp.
◆ stlStrToStlWStr()
|
inline |
Definition at line 115 of file stringFuncs.h.
References nullifyMarker().

◆ stlWStrToStlStr()
|
inline |
Definition at line 108 of file stringFuncs.h.
◆ strAppend()
| void AtomProbe::strAppend | ( | std::string | s, |
| const T & | a | ||
| ) |
Definition at line 41 of file stringFuncs.h.
References convertFileStringToCanonical(), convertFileStringToNative(), digitString(), genColString(), lowercase(), onlyDir(), onlyFilename(), parseColString(), splitStrsRef(), stream_cast(), stripChars(), stripWhite(), stripZeroEntries(), and uppercase().
Referenced by AtomProbe::RangeFile::isSelfConsistent().
◆ stream_cast()
| bool AtomProbe::stream_cast | ( | T1 & | result, |
| const T2 & | obj | ||
| ) |
Template function to cast and object to another by the stringstream.
Definition at line 32 of file stringFuncs.h.
Referenced by AtomProbe::RangeFile::decomposeIonNames(), AtomProbe::RangeFile::detectFileType(), digitString(), AtomProbe::LibVersion::getVersionStr(), AtomProbe::Mesh::loadGmshMesh(), loadTapsimBinFile(), loadTextData(), main(), AtomProbe::MultiRange::open(), AtomProbe::RangeFile::open(), AtomProbe::Point3D::parse(), parseCompositionData(), rangeToStr(), AtomProbe::RangeFile::rangeTypeString(), readPosapOps(), saveTapsimBin(), strAppend(), XMLGetNextElemAttrib(), and XMLHelpGetProp().
◆ strhas()
| bool AtomProbe::strhas | ( | const char * | cpTest, |
| const char * | cpPossible | ||
| ) |
Definition at line 1485 of file dataFiles.cpp.
Referenced by loadTextData().
◆ stripChars()
| std::string AtomProbe::stripChars | ( | const std::string & | Str, |
| const char * | chars | ||
| ) |
Definition at line 137 of file stringFuncs.cpp.
Referenced by AtomProbe::RangeFile::rangeTypeString(), strAppend(), and stripWhite().
◆ stripWhite()
| std::string AtomProbe::stripWhite | ( | const std::string & | str | ) |
Strip whitespace, (eg tab,space) from either side of a string.
Definition at line 132 of file stringFuncs.cpp.
References stripChars().
Referenced by AtomProbe::RangeFile::detectFileType(), main(), AtomProbe::Point3D::parse(), parseCompositionData(), AtomProbe::RangeFile::rangeTypeString(), readPosapOps(), and strAppend().

◆ stripZeroEntries()
| void AtomProbe::stripZeroEntries | ( | std::vector< std::string > & | s | ) |
Definition at line 155 of file stringFuncs.cpp.
Referenced by AtomProbe::RangeFile::detectFileType(), loadTextData(), main(), parseCompositionData(), AtomProbe::RangeFile::rangeTypeString(), readPosapOps(), and strAppend().
◆ sumVoxels()
| void AtomProbe::sumVoxels | ( | const Voxels< T > & | src, |
| U & | counter | ||
| ) |
Use one counting type to sum counts in a voxel of given type.
Definition at line 376 of file voxels.h.
References AtomProbe::Voxels< T >::getData(), AtomProbe::Voxels< T >::getSize(), and AtomProbe::Voxels< T >::size().
◆ tabs()
|
inline |
Definition at line 99 of file stringFuncs.h.
Referenced by AtomProbe::MultiRange::write().
◆ tolEqual()
| bool AtomProbe::tolEqual | ( | const T & | a, |
| const T & | b, | ||
| const T & | f | ||
| ) |
Test for equality within tolerance f (||a-b|| < f)
Definition at line 59 of file misc.h.
Referenced by AtomProbe::ModifiedFocusSphericProjection::scaleUp().
◆ transposeVector()
| void AtomProbe::transposeVector | ( | std::vector< std::vector< T > > & | v | ) |
◆ triIsDegenerate()
Definition at line 266 of file misc.cpp.
References AtomProbe::Point3D::mag().
Referenced by dotProduct(), and GetReducedHullPts().
◆ ucharToHexStr()
| void AtomProbe::ucharToHexStr | ( | unsigned char | c, |
| std::string & | s | ||
| ) |
Definition at line 68 of file stringFuncs.cpp.
Referenced by genColString().
◆ uppercase()
| std::string AtomProbe::uppercase | ( | std::string | s | ) |
Return a uppercase version for a given string.
Referenced by strAppend().
◆ vectorMultiErase()
| void AtomProbe::vectorMultiErase | ( | std::vector< T > & | vec, |
| const std::vector< bool > & | wantKill | ||
| ) |
Remove elements from the vector, without preserving order.
Definition at line 112 of file misc.h.
Referenced by computeIonDistAdjacency(), AtomProbe::RangeFile::decomposeIonNames(), AtomProbe::RangeFile::eraseIon(), filterBySolutionPPM(), filterPeakNeedBiggerObs(), findOverlaps(), AtomProbe::AbundanceData::generateIsotopeDist(), AtomProbe::RangeFile::makeSelfConsistent(), and FixedStack< T >::~FixedStack().
◆ vectorPointDir()
| unsigned int AtomProbe::vectorPointDir | ( | const Point3D & | pA, |
| const Point3D & | pB, | ||
| const Point3D & | vC, | ||
| const Point3D & | vD | ||
| ) |
Check which way vectors attached to two 3D points "point",.
Two vectors may point "together", /__\ "apart" __/ or "In common" /__/ or __\
Definition at line 127 of file misc.cpp.
References ASSERT, POINTDIR_APART, POINTDIR_IN_COMMON, and POINTDIR_TOGETHER.
Referenced by distanceToSegment(), and meanAndStdev().
◆ voigtProfile() [1/2]
| void AtomProbe::voigtProfile | ( | const std::vector< double > & | x, |
| double | sigma, | ||
| double | gamma, | ||
| double | mu, | ||
| double | amp, | ||
| std::vector< double > & | y | ||
| ) |
Generate a shifted voigt profile.
Definition at line 486 of file fitting.cpp.
References AtomProbe::DATA_FIT::x.
Referenced by fitVoigtFunc(), lsq(), and main().
◆ voigtProfile() [2/2]
| void AtomProbe::voigtProfile | ( | const vector< double > & | x, |
| double | sigma, | ||
| double | gamma, | ||
| vector< double > & | y | ||
| ) |
Definition at line 449 of file fitting.cpp.
References M_PI.
◆ weightedMean()
| T AtomProbe::weightedMean | ( | const std::vector< T > & | values, |
| const std::vector< T > & | weight | ||
| ) |
Definition at line 52 of file misc.h.
Referenced by fitDoniachSunjic(), fitExpNorm(), fitLikeLogGaussian(), fitVoigt(), and AtomProbe::AbundanceData::generateGroupedIsotopeDist().
◆ XMLFreeDoc()
| void AtomProbe::XMLFreeDoc | ( | void * | data | ) |
Free a xmlDoc pointer. For use in conjunction with std::unique_ptr for auto-deallocation.
Definition at line 23 of file XMLHelper.cpp.
Referenced by AtomProbe::MultiRange::open().
◆ XMLGetNextElemAttrib()
| bool AtomProbe::XMLGetNextElemAttrib | ( | xmlNodePtr & | nodePtr, |
| T & | v, | ||
| const char * | nodeName, | ||
| const char * | attrib | ||
| ) |
Grab the specified attribute from the next element, then stream_cast() it into the passed-in object. Returns true on success, false on error.
Definition at line 112 of file XMLHelper.h.
References stream_cast(), and XMLHelpFwdToElem().
◆ XMLHelpFwdNotElem()
| unsigned int AtomProbe::XMLHelpFwdNotElem | ( | xmlNodePtr & | node, |
| const char * | nodeName | ||
| ) |
Definition at line 53 of file XMLHelper.cpp.
◆ XMLHelpFwdToElem()
| unsigned int AtomProbe::XMLHelpFwdToElem | ( | xmlNodePtr & | node, |
| const char * | nodeName | ||
| ) |
Definition at line 41 of file XMLHelper.cpp.
Referenced by AtomProbe::AbundanceData::open(), AtomProbe::MultiRange::open(), and XMLGetNextElemAttrib().
◆ XMLHelpFwdToList()
| unsigned int AtomProbe::XMLHelpFwdToList | ( | xmlNodePtr & | node, |
| const std::vector< std::string > & | nodeList | ||
| ) |
◆ XMLHelpGetProp()
| unsigned int AtomProbe::XMLHelpGetProp | ( | T & | prop, |
| xmlNodePtr | node, | ||
| std::string | propName | ||
| ) |
Definition at line 87 of file XMLHelper.h.
References PROP_BAD_ATT, PROP_PARSE_ERR, and stream_cast().
Referenced by AtomProbe::AbundanceData::open(), and AtomProbe::MultiRange::open().

◆ XMLHelpGetText() [1/2]
| string AtomProbe::XMLHelpGetText | ( | xmlNodePtr | node | ) |
Definition at line 64 of file XMLHelper.cpp.
References XMLHelpNextType().

◆ XMLHelpGetText() [2/2]
| std::string AtomProbe::XMLHelpGetText | ( | xmlNodePtr & | node | ) |
◆ XMLHelpNextType()
| unsigned int AtomProbe::XMLHelpNextType | ( | xmlNodePtr & | node, |
| int | nodeType | ||
| ) |
Definition at line 28 of file XMLHelper.cpp.
Referenced by XMLHelpGetText().
◆ zechConfidenceLimits()
| bool AtomProbe::zechConfidenceLimits | ( | float | lambdaBack, |
| unsigned int | observation, | ||
| float | alpha, | ||
| float & | estimate | ||
| ) |
Provides a best estimate for true signal when true signal, background.
Nuclear Instruments and Methods in Physics Research A, 1989 608-610 "Upper limits in Experiments with Background or Measurement Errors" This is numerically unstable for large counts A similar problem is tackled by Bityukov, arxiv:hep-ex/0108020v1 Equation 3.5, but is less numerically stable
lambdaBack : background rate observation : Observed signal+background alpha : confidence parameter, (0,1) estimate : estimate for signal at confidence bound, specified by alpha Returns true if calculation succeeds, false otherwise. Note :this involves a root-finding approach, so may be slow
NOTE: may not be numerically stable for high lambdaback/observation, or lambdaback>>observation. This may cause GSL to error out.
Definition at line 406 of file confidence.cpp.
References AtomProbe::ZECH_ROOT::alpha, ASSERT, AtomProbe::ZECH_ROOT::lambdaBack, AtomProbe::ZECH_ROOT::observation, and zechRoot().
Referenced by poissonConfidenceObservation(), and zechCorrect().
◆ zechRoot()
| double AtomProbe::zechRoot | ( | double | sigGuess, |
| void * | params | ||
| ) |
Definition at line 386 of file confidence.cpp.
References AtomProbe::ZECH_ROOT::alpha, AtomProbe::ZECH_ROOT::lambdaBack, and AtomProbe::ZECH_ROOT::observation.
Referenced by zechConfidenceLimits().
Variable Documentation
◆ a2iTriangleConnectionTable
| int AtomProbe::a2iTriangleConnectionTable[256][16] |
Definition at line 194 of file isoSurface.cpp.
◆ ABUNDANCE_ERROR
| const char* AtomProbe::ABUNDANCE_ERROR[] |
Definition at line 49 of file abundance.cpp.
◆ aiCubeEdgeFlags
| int AtomProbe::aiCubeEdgeFlags[256] |
Definition at line 166 of file isoSurface.cpp.
◆ ATO_ERR_STRINGS
| const char * AtomProbe::ATO_ERR_STRINGS |
Human readable error messages for use with ATO reader return values.
Definition at line 73 of file dataFiles.cpp.
◆ edgeRemap
| int AtomProbe::edgeRemap[12] |
Definition at line 488 of file isoSurface.cpp.
◆ ELEM_FOUR_NODE_TETRAHEDRON
| const unsigned int AtomProbe::ELEM_FOUR_NODE_TETRAHEDRON =4 |
Definition at line 38 of file mesh.h.
Referenced by AtomProbe::Mesh::loadGmshMesh(), and AtomProbe::Mesh::saveGmshMesh().
◆ ELEM_SINGLE_NODE_POINT
| const unsigned int AtomProbe::ELEM_SINGLE_NODE_POINT =15 |
Definition at line 35 of file mesh.h.
Referenced by AtomProbe::Mesh::loadGmshMesh(), and AtomProbe::Mesh::saveGmshMesh().
◆ ELEM_THREE_NODE_TRIANGLE
| const unsigned int AtomProbe::ELEM_THREE_NODE_TRIANGLE =2 |
Definition at line 37 of file mesh.h.
Referenced by AtomProbe::Mesh::loadGmshMesh(), and AtomProbe::Mesh::saveGmshMesh().
◆ ELEM_TWO_NODE_LINE
| const unsigned int AtomProbe::ELEM_TWO_NODE_LINE =1 |
Definition at line 36 of file mesh.h.
Referenced by AtomProbe::Mesh::loadGmshMesh(), and AtomProbe::Mesh::saveGmshMesh().
◆ elementList
| const char* AtomProbe::elementList[] |
Definition at line 63 of file ranges.cpp.
◆ EPOS_RECORD_SIZE
| const size_t AtomProbe::EPOS_RECORD_SIZE = 11*4 |
Definition at line 144 of file ionHit.h.
Referenced by chunkLoadEposFile(), and loadEposFile().
◆ hardAssert
| bool AtomProbe::hardAssert =true |
Definition at line 23 of file aptAssert.cpp.
Referenced by getHardAssert().
◆ HULL_GRAB_SIZE
| const unsigned int AtomProbe::HULL_GRAB_SIZE =4096 |
Definition at line 42 of file convexHull.cpp.
◆ libVersion
| LibVersion AtomProbe::libVersion |
Definition at line 28 of file atomprobe.cpp.
Referenced by AtomProbe::RandNumGen::getRng().
◆ MAX_LINE_SIZE
| const size_t AtomProbe::MAX_LINE_SIZE = 16536 |
Definition at line 51 of file ranges.cpp.
Referenced by AtomProbe::RangeFile::open(), and AtomProbe::RangeFile::rangeTypeString().
◆ MAX_RANGEFILE_SIZE
| const size_t AtomProbe::MAX_RANGEFILE_SIZE = 50*1024*1024 |
Definition at line 53 of file ranges.cpp.
◆ maximumLinearTable
| const size_t AtomProbe::maximumLinearTable[] |
Definition at line 39 of file lfsr.cpp.
Referenced by AtomProbe::LinearFeedbackShiftReg::verifyTable().
◆ MESH_LOAD_ERRS
| const char * AtomProbe::MESH_LOAD_ERRS |
◆ MULTIRANGE_FORMAT_VERSION
| const char AtomProbe::MULTIRANGE_FORMAT_VERSION[] = "0.0.1" |
Definition at line 46 of file multiRange.cpp.
◆ NUM_ELEMENTS
◆ OPS_ENUM_ERRSTRINGS
| const char * AtomProbe::OPS_ENUM_ERRSTRINGS |
Definition at line 776 of file dataFiles.cpp.
◆ PROGRESS_REDUCE
| const size_t AtomProbe::PROGRESS_REDUCE =500 |
Definition at line 61 of file mesh.cpp.
Referenced by AtomProbe::K3DTreeExact::build(), computeConvexHull(), and AtomProbe::Mesh::pointsInside().
◆ qhullInited
| bool AtomProbe::qhullInited =false |
Definition at line 39 of file convexHull.cpp.
◆ randGen
| RandNumGen AtomProbe::randGen |
Definition at line 29 of file atomprobe.cpp.
Referenced by AtomProbe::RandNumGen::getRng(), leastSquaresOverlapSolve(), loadATOFile(), loadPosFile(), main(), poissonConfidenceObservation(), and sampleIons().
◆ RANGE_EXTS
| const char* AtomProbe::RANGE_EXTS[] |
Definition at line 55 of file ranges.cpp.
◆ RECORDREAD_ERR_STRINGS
| const char * AtomProbe::RECORDREAD_ERR_STRINGS |
Definition at line 82 of file dataFiles.cpp.
Referenced by main().
◆ VERTEX_OFFSET
| const unsigned int AtomProbe::VERTEX_OFFSET[8][3] |
Definition at line 462 of file isoSurface.cpp.
 1.8.13
1.8.13